Abstract
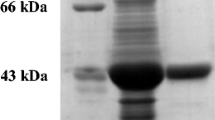
Nitrilases are of commercial interest in the selective synthesis of carboxylic acids from nitriles. Nitrilase induction was achieved here in three bacterial strains through the incorporation of a previously unrecognised and inexpensive nitrilase inducer, dimethylformamide (DMF), during cultivation of two Rhodococcus rhodochrous strains (ATCC BAA-870 and PPPPB BD-1780), as well as a closely related organism (Pimelobacter simplex PPPPB BD-1781). Benzonitrile, a known nitrilase inducer, was ineffective in these strains. Biocatalytic product profiling, enzyme inhibition studies and protein sequencing were performed to distinguish the nitrilase activity from that of sequential nitrile hydratase-amidase activity. The expressed enzyme, a 40-kDa protein with high sequence similarity to nitrilase protein Uniprot Q-03217, hydrolyzed 3-cyanopyridine to produce nicotinic acid exclusively in strains BD-1780 and BD-1781. These strains were capable of synthesising both the vitamin nicotinic acid as well as β-amino acids, a compound class of pharmaceutical interest. The induced nitrilase demonstrated high enantioselectivity (> 99%) in the hydrolysis of 3-amino-3-phenylpropanenitrile to the corresponding carboxylic acid.







Similar content being viewed by others
References
Ahmed K, Kumar M, MS KCG, Shaik TB (2011) Bioconversion of acrylonitrile to acrylic acid by Rhodococcus ruber strain AKSH-84. J Microbiol Biotechnol 21:37–42
Almatawah QA, Cramp R, Cowan DA (1999) Characterization of an inducible nitrilase from a thermophilic bacillus. Extremophiles 3:283–291
Badoei-Dalfard A, Karami Z, Ramezani-pour N (2016) Bench scale production of nicotinic acid using a newly isolated Stenotrophomonas maltophilia AC21 producing highly-inducible and versatile nitrilase. J Mol Catal B Enzym 133:S552–S559
Bauer R, Knackmuss H-J, Stolz A (1998) Enantioselective hydration of 2-arylpropionitriles by a nitrile hydratase from Agrobacterium tumefaciens strain d 3. Appl Microbiol Biotechnol 49:89–95
Binfeng L, Jinhuan S, Tao J (2011) Enzyme and process development for production of nicotinamide. Org Process Res Dev 15:291–293
Borsook H, Davenport HW, Jeffreys CEP, Warner RC (1937) The oxidation of ascorbic acid and its reduction in vitro and in vivo. J Biol Chem 117:237–279
Brady D, Beeton A, Kgaje C, Zeevaart J, van Rantwijk F, Sheldon RA (2004) Characterisation of nitrilase and nitrile hydratase biocatalytic systems. Appl Microbiol Biotechnol 64:76–85
Brenner C (2002) Catalysis in the nitrilase superfamily. Curr Opin Struct Biol 12:775–782
Chhiba VP, Bode ML, Mathiba K, Kwezi W, Brady D (2012) Enantiomeric biocatalytic hydrolysis of β-aminonitriles to β-aminoamides using Rhodococcus rhodochrous ATCC BAA-870. J Mol Catal B Enzym 76:68–74
Chhiba VP, Bode M, Mathiba K, Brady D (2014) Enzymatic stereoselective synthesis of β-amino acids. In: Riva S, Fessner WD (eds) Cascade biocatalysis: integrating stereoselective and environmentally friendly reactions. Wiley-VCH, Germany, pp 297–313
Cramp RA, Cowan DA (1999) Molecular characterization of a novel thermophilic nitrile hydratase. Biochem Biophys Acta 1431:249–260
Endo T, Watanabe I (1989) Nitrile hydratase of Rhodococcus sp. N774 purification and amino acid sequences. FEBS Lett 243:61–64
Fang S, An X, Liu H, Cheng Y, Hou N, Feng L, Huang X, Li C (2015) Enzymatic degradation of aliphatic nitriles by Rhodococcus rhodochrous BX2, a versatile nitrile-degrading bacterium. Bioresour Technol 185:28–34
Fernandes BCM, Mateo C, Kiziak C, Chmura A, Wacker J, van Rantwijk F, Stolz A, Sheldon RA (2006) Nitrile hydratase activity of a recombinant nitrilase. Adv Synth Catal 348:2597–2603
Goncalves ER, Hara H, Miyazawa D, Davies JE, Eltis LP, Mohn WW (2006) Transcriptomic assessment of isozymes in the biphenyl pathway of Rhodococcus sp. strain RHA1. Appl Environ Microbiol 72:6183–6193
Gong J-S, Lu Z-M, Li H, Shi JS, Zhou Z-M, Xu Z-H (2012) Nitrilases in nitrile biocatalysis: recent progress and forthcoming research. Microb Cell Factories 11:142
Gradley ML, Deverson CJF, Knowles CJ (1994) Asymmetric hydrolysis of R-(-)-S (+)-2-methylbutyronitrile by Rhodococcus rhodochrous NCIMB 11216. Arch Microbiol 161:246–251
Gregoriou M, Brown PR (1979) Inhibition of the aliphatic amidase from Pseudomonas aeruginosa by urea and related compounds. Eur J Biochem 96:101–108
Harper DB (1977) Microbial metabolism of aromatic nitriles: enzymology of aromatic nitriles: enzymology of C-N cleavage by Nocardia sp. (Rhodococcus group) N. C. I. B. 11216. Biochem J 165:309–319
He Y-C, Wu Y-D, Pan X-H, Ma C-L (2014) Biosynthesis of terephthalic acid, isophthalic acid and their derivatives from the corresponding dinitriles by tetrachloroterephthalonitrile-induced Rhodococcus sp. Biotechnol Lett 36:341–347
Herai S, Hashimoto Y, Higashibata H, Maseda H, Ikeda H, Omura S, Kobayashi M (2004) Hyper-inducible expression system for Streptomycetes. Proc Natl Acad Sci 101:14031–14035
Kaul P, Banerjee UC (2008) Predicting enzyme behaviour in nonconventional media: correlating nitrilase function with solvent properties. J Ind Microbiol Biotechnol 35:713–720
Kinfe HH, Chhiba V, Frederick J, Bode ML, Mathiba K, Steenkamp PA, Brady D (2009) Enantioselective hydrolysis of β-hydroxy nitriles using the whole cell biocatalyst Rhodococcus rhodochrous ATCC BAA-870. J Mol Catal B Enzym 59:231–236
Kobayashi M, Nagasawa T, Yamada H (1989) Nitrilase of Rhodococcus rhodochrous J1. Eur J Biochem 182:349–356
Kobayashi M, Goda M, Shimizu S (1998) The catalytic mechanism of amidase also involves nitrile hydrolysis. FEBS Lett 439:325–328
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Layh N, Parratt J, Willetts A (1998) Characterization and partial purification of an enantioselective arylacetonitrilase from Pseudomonas fluorescens DSM 7155. J Mol Catal B Enzym 5:476–474
Leonova TE, Astaurova OB, Ryabchenko LE, Yanenko AS (2000) Nitrile hydratase of Rhodococcus: optimization of synthesis in cells and industrial applications for acrylamide production. Appl Biochem Biotechnol 88:231–241
Lévi-Schil S, Soubrier F, Crutz-Le Coq A-M, Faucher D, Crouzet J, Pétré D (1995) Aliphatic nitrilase from a soil-isolated Comomonas testosteroni sp.: gene cloning and overexpression, purification and primary structure. Gene 161:15–20
Martínková L, Uhnáková B, Pátek M, Nešvera J, Křen V (2009) Biodegradation potential of the genus Rhodococcus. Environ Int 35:162–177
Martínková L, Rucká L, Nešvera J, Pátek M (2017) Recent advances and challenges in the heterologous production of microbial nitrilases for biocatalytic applications. World J Microbiol Biotechnol 33:8
Mateo C, Chmura A, Rustler S, van Rantwijk F, Stolz A, Sheldon RA (2006) Synthesis of enantiomerically pure (S)-mandelic acid using an oxynitrilase-nitrilase bienzymatic cascade: a nitrilase surprisingly shows nitrile hydratase activity. Tetrahedron Asymmetry 17:320–323
Miller JM, Conn EE (1980) Metabolism of hydrogen cyanide by higher plants. Physiol Plant 65:1199–1202
Nagasawa T, Ryuno K, Yamada H (1986) Nitrile hydratase of Brevibacterium R-312 purification and characterisation. Biochem Biophys Res Commun 139:1304–1312
Nagasawa T, Nakamura T, Yamada H (1990) ε-Caprolactam, a new powerful inducer for the formation of Rhodococcus rhodochrous J1 nitrilase. Arch Microbiol 155:13–17
Nagasawa T, Takeuchi K, Yamda H (1991) Characterization of a new cobalt-containing nitrile hydratase purified from urea-induced cells of Rhodococcus rhodochrous J1. Eur J Biochem 196:581–589
Nagasawa T, Weiser M, Nakamura T, Iwahara H, Yoshida T, Gekko K (2000) Nitrilase or Rhodococcus rhodochrous J1. Conversion into the active form by subunit association. Eur J Biochem 267:138–144
Nageshwar YVD, Sheelu G, Shambu RR, Muluka H, Mehdi N, Malik MS, Kamal A (2011) Optimization of nitrilase production from Alcaligenes faecalis MTCC 10757 (IICT-A3): effect of inducers on substrate specificity. Bioprocess Biosyst Eng 34:515–523
Pertsovich SI, Guranda DT, Podchernyaev DA, Yanenko AS, Svedas VK (2005) Aliphatic amidase from Rhodococcus rhodochrous M8 is related to the nitrilase/cyanide hydratase family. Biochem Mosc 70:1280–1287
Petříčková A, Veselá AB, Kaplan O, Kubáč D, Uhnáková B, Malandra A, Felsberg J, Rinágelová A, Weyrauch P, Křen V, Bezouška K, Martínková L (2012) Purification and characterization of heterologously expressed nitrilases from filamentous fungi. Appl Microbiol Biotechnol 93:1553–1561
Prasad S, Misra A, Jangir VP, Awasthi A, Raj J, Bhalla TC (2007) A propionitrile-induced nitrilase of Rhodococcus sp. NDB 1165 and its application in nicotinic acid synthesis. World J Microbiol Biotechnol 23:345–353
Prasad S, Raj J, Bhalla TC (2009) Purification of a hyperactive nitrile hydratase from resting cells of Rhodococus rhodochrous PA-34. Indian J Microbiol 49:237–242
Precigou S, Wieser M, Pommares P, Goulas P, Duran R (2004) Rhodococcus pyridinovorans MW3, a bacterium producing a nitrile hydratase. Biotechnol Lett 26:1379–1384
Rapheeha OK, Roux-van der Merwe MP, Badenhorst J, Chhiba V, Bode ML, Mathiba K, Brady D (2017) Hydrolysis of nitriles by soil bacteria: variation with soil origin. J Appl Microbiol 122:686–697
Robinson WG, Hook RH (1964) Ricinine nitrilase: I. Reaction producer and substrate specificity. J Biol Chem 239:4257–4262
Sharma NN, Sharma M, Bhalla TC (2011) An improved nitrilase-mediated bioprocess for synthesis of nicotinic acid from 3-cyanopyridine with hyperinduced Nocardia globerula NHB-2. J Ind Microbiol Biotechnol 38:1235–1243
Stalker DM, Malyj LB, McBride KE (1988) Properties of a nitrilase specific for the herbicide bromoxynil and corresponding nucleotide sequence analysis of the bxn gene. J Biol Chem 263:6310–6314
Stevenson DE, Feng R, Dumas F, Groleau D, Mihoc A, Storer AC (1992) Mechanistic and structural studies on Rhodococcus ATCC 39484 nitrilase. Biotechnol Appl Biochem 15:283–302
Tani Y, Kurihara M, Nishise H (1989) Characterization of nitrile hydratase and amidase, which are responsible for the conversion of dinitriles to mononitriles, from Corynebacterium sp. Agric Biol Chem 53:3151–3158
Tauber MM, Cavaco-Paulo A, Robra K, Gubitz GM (2000) Nitrile hydratase and amidase from Rhodococcus rhodochrous hydrolyze acrylic fibers and granular polyacrylonitriles. Appl Environ Microbiol 66:1634–1638
Thalenfeld B, Grossowicz N (1976) Regulatory properties of an inducible aliphatic amidase in a thermophilic Bacillus. J Gen Microbiol 94:131–141
Thuku RN, Brady D, Benedik MJ, Sewell BT (2009) Microbial nitrilases: versatile, spiral forming enzymes. J Appl Microbiol 106:703–727
Vaudel M, Burkhart JM, Zahedi RP, Oveland E, Berven FS, Sickmann A, Martens L, Barsnes H (2015) PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat Biotechnol 33:22–24
Vejvoda V, Kubác D, Davidová A, Kaplan O, Śulc M, Śveda O, Chaloupková R, Martínkova L (2010) Purification and characterization of nitrilase from Fusarium solani IMI96840. Process Biochem 45:1115–1120
Yamada H, Kobayashi M (1996) Nitrile hydratase and its application to industrial production of acrylamide. Biosci Biotechnol Biochem 60:1391–1400
Yusuf F, Chaubey A, Jamwal U, Parshad R (2013) A new isolate from Fusarium proliferatum (AUF-2) for efficient nitrilase production. Appl Biochem Biotechnol 171:1022–1031
Zangi R, Zhou R, Berne BJ (2009) Urea’s action on hydrophobic interactions. J Am Chem Soc 131:1535–1541
Zhang ZJ, Xu JH, He YC, Ouyang LM, Liu YY (2011) Cloning and biochemical properties of a highly thermostable and enantioselective nitrilase from Alcaligenes sp. ECU0401 and its potential for (R)-(-)-mandelic acid production. Bioprocess Biosyst Eng 34:315–322
Zheng JG, Chen J, Liu ZQ, Wu MH, Xing LY, Shen YC (2008) Isolation, identification and characterization of Bacillus subtilis ZJB-063, a versatile nitrile-converting bacterium. Appl Microbiol Biotechnol 77:985–993
Websites
Merops-the peptidase database 2017, accessed 20th March 2017, <https://www.ebi.ac.uk/merops/cgi-bin/speccards?sp=sp034913;type=peptidase>
Acknowledgements
We would like to thank the following people at the University of the Witwatersrand: Prof. Joseph Michael; Prof. Charles De Koning; and Dr. Amanda Rousseau. At the CSIR, we would like to extend thanks to Mr. Sipho Mamputhu (for assistance with LC-MSMS protein sequencing) as well as Dr. Priyen Pillay, Dr. Lusisiwe Kwezi, Dr. Ofense Poe, Advaita Singh, Sibongile Mtimka, Dr. Daria Rutkovska, Dr. Kevin Wellington and Dr. Tsepo Tsekoa.
Funding
Support was received from the Department of Science and Technology Biocatalysis Initiative (Grant 0175/2013).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 649 kb)
Rights and permissions
About this article
Cite this article
Chhiba-Govindjee, V.P., Mathiba, K., van der Westhuyzen, C.W. et al. Dimethylformamide is a novel nitrilase inducer in Rhodococcus rhodochrous. Appl Microbiol Biotechnol 102, 10055–10065 (2018). https://doi.org/10.1007/s00253-018-9367-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9367-9