Abstract
Even though traditional breeding of perennial fruit trees such as apple and pear has resulted in high performing cultivars in the past, it is a very lengthy and costly process that is unable to keep up with the increasing demands for improved yield, resistance and fruit quality posed by the growing world population and the rapidly changing climate. In the last decade, significant research advances have been made that can revolutionize pome fruit breeding to meet current needs, including the sequencing of apple and pear genomes, the increased understanding of associations between gene(s) and traits of interest, and the advancement in genetic engineering tools. In particular the emergence of genome-editing tools such as the CRISPR/Cas9 technology can significantly improve the speed and accuracy of pome fruit breeding programs. This chapter reviews the progress, opportunities and challenges of genome editing tools in apple and pear, and discusses the genetic basis of several important breeding goals to find possible targets for new gene-editing applications.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
1 Introduction
The term “pome” refers to an accessory fruit produced by temperate tree species belonging to the maleae tribe of the Rosaceae family. The best-known and economically most important pome fruits are apple and pear. In 2020, apples were among the five most produced fruit crops worldwide together with watermelons, bananas, oranges and grapes with 88.3 million metric tons [1]. Both apples and pears are commercially grown in over 50 countries with China being the top producer for both fruit crops [2]. Due to their economic importance and the wide geographic distribution of their production, ongoing breeding programs are present all over the world aiming to improve various traits including higher yield, increased disease resistance, and improved fruit quality [3,4,5].
Pome fruit breeding is an expensive and lengthy process due to biological characteristics typical of woody tree species, namely a long juvenile period (5–7 years), self-incompatibility, high heterozygosity, a limited available gene pool for new traits, and a large genome size and chromosome number [6, 7]. In a classical breeding scheme, selected parents are intercrossed to create hybrid seedling populations, consisting of a pool of unique genotypes, followed by a strict selection of the best performing progeny clone. Due to the limited genetic variation present in advanced breeding material and established cultivars, it is often necessary to use wild or semi-wild gene pools as sources for the introduction of new traits. Pre-breeding to obtain suitable parents from these (semi-)wild gene pools that can be crossed with elite germplasm takes decades due to the long life cycle and the need for repeated cycles. More specifically, the introgression of desired alleles originating from donor varieties with a minimum of linkage drag and concomitant selection for various other traits, requires multiple generations of hybridizations followed by selection, and thus significantly adds to the breeding time [7]. Alternatively, non-GMO breeding techniques that are used include interspecific hybridization, induced mutagenesis and polyploidization. Also, a wide range of genetic and molecular tools have been used to improve the efficiency and selection accuracy of pome fruit breeding programs such as genetic linkage maps, molecular markers and whole genome sequencing [8,9,10,11]. Each of these techniques have their own advantages and drawbacks, but many of the limitations remain similar to traditional breeding.
Recently, the introduction of genome editing techniques such as CRISPR/CAS has provided efficient ways to introduce precise mutations in plants. These techniques are especially beneficial in clonally propagated fruit tree species since they can generate improved breeding outcomes compared to conventional techniques without the extensive backcrossing and associated linkage drag that is necessary when introgressing new traits in established cultivars [12]. These genome editing techniques have the potential to greatly accelerate and improve the breeding process of woody fruit tree species, even though important challenges and limitations still need to be overcome to allow their broad scale application.
This chapter reviews the recent progress in the application of genome editing techniques in apple and pear, as well as the specific opportunities and challenges. We also outline and discuss important genes underlying economically interesting traits which could be used as valuable targets for gene-editing.
2 Genome Editing Technologies in Pome Fruit Trees
Genome editing refers to the use of genetic transformation techniques that can be used to precisely edit the plant genome [13]. There are three main genome editing approaches that are all based on the use of engineered nucleases, namely zinc finger nucleases (ZFNs), transcription activator-like effector-based nucleases (TALEN) and clustered regularly interspaced short palindromic repeats associated nucleases (CRIPSR/cas) [3]. These nucleases can be designed to bind a specific target DNA sequence in the genome of the plant where they induce a double-strand break (DSB) which is subsequently repaired by one of the two following processes, homology directed DNA break repair (HDR) or non-homologous end joining (NHEJ) [14, 15]. Plant transformation is generally performed using Agrobacterium tumefaciens followed by regeneration of transgenic tissue in vitro. This paragraph describes the current use, adaptations and limitations of these three genome editing techniques in pome fruit breeding.
2.1 Zinc Finger Nucleases (ZFNs)
Zinc finger nucleases were one of the first editing technologies [16]. They are artificial enzymes generated by fusing a zinc-finger DNA-binding domain to a nonspecific DNA-cleavage domain of the Fok I endonuclease enzyme [13, 17]. A pair of custom designed ZFNs bind to the DNA at the target location and together form an active dimer nuclease complex [18]. ZFNs have been successfully used for targeted mutagenesis in many species including Arabidopsis [19, 20], soybean [21], rice [22], and populus [23]. To our knowledge, only a single successful instance of ZFN application in pome fruit has been published [13]. The authors validated the use of ZFNs in apple and fig using a visual transgenic repair assay based on activation of a mutated uidA gene, which encodes the GUS reporter protein. The overall efficiency in apple was around 10% and the authors concluded that the genome editing approach was suitable for application in fruit tree species. However, although the technique has been around for almost three decades, the adoption of ZFNs in plant breeding is limited mainly due to their low efficiency, the complex construction of the zinc finger region that interacts with DNA and severe off-target effects [24,25,26,27].
2.2 Transcription Activator-Like Effector Nucleases (TALENs)
Another genome editing approach is based on the family of proteins known as transcription activator-like effectors (TALEs) which are produced in bacteria including plant pathogenic Xanthomonas species. These effector proteins have a DNA binding domain which is linked with a catalytic domain of an endonuclease like Fok I [18, 25]. TALE nucleases (TALENs) with desired specificities can be created through modification of the DNA binding domain to target specific locations in the genome. The nucleases there act like molecular scissors that produce DSBs which are repaired with the cellular repair mechanisms leading to deletions, insertions, replacements or rearrangements [18, 25].
TALEN-mediated genome editing has been successfully applied in rice [28, 29], wheat [30], maize [31], and sugarcane [32, 33] among others. However, to our knowledge, no application of TALEN-based transformation has been applied in apple or pear or even any other woody fruit tree species such as citrus or prunus. Although TALENs have some advantages over ZFNs, such as lower toxicity and somewhat simplified construction, the CRISPR/Cas approach has quickly surpassed both ZFNs and TALENs as method of choice in plant genome editing [26, 27].
2.3 CRISPR/Cas Systems
Currently, clustered regularly interspaced short palindromic repeats (CRISPR)-based genome editing has become the most reliable and cost-effective approach in plant research. Unlike ZFN and TALEN, the DNA-recognition, based on RNA-DNA interactions, is faster, cheaper and generally more efficient [17, 34]. In plants, CRISPR/Cas 9 is the most used system and consists of a Cas 9 nuclease and a single guide RNA which replaces the original CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracr RNA). The gRNA contains a unique sequence of 20 bp which preceeds a protospacer adjacent motif (PAM). This gRNA binds the Cas 9 nuclease and directs it to a complementary target sequence on the genomic DNA. The two nuclease domains of the Cas 9 protein (RuvC and HNH) will then cleave the target sequence at three nucleotides upstream of the PAM site, leaving predominantly blunt ends [26, 35].
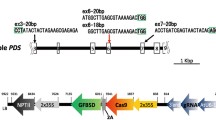
The CRISPR/Cas9 method has been widely adopted in plant research and has been used in many crops including pome fruit species. It was first used in apple to modify an apple phytoene desaturase (PDS) gene which encodes an essential plant carotenoid biosynthetic enzyme required for chlorophyll biosynthesis. Knock-out of this gene leads to an albino phenotype making it an easy visual marker [34, 36]. CRISPR/Cas9 was also used to reduce susceptibility to fire blight in apple [37], and induce early flowering in apple and pear [38]. It has also been applied in wild apple to target the MsPDS gene [39]. To date, nearly all applications in pome fruit use the CRISPR/Cas9 system, however alternative CRISPR systems can be applied as well [40]. One example is the CRISPR/Cas12 system, previously known as Cpf1, which also belongs to the class 2 CRISPR systems, but lacks the HNH domain and generates a staggered cut with a 5 nt 5′ overhang [41]. CRISPR/Cas12 has been successfully applied in other woody, perennial tree species such as poplar [42] and citrus [43]. Alternative CRISPR/Cas systems may be more suitable in specific situations, for example the CRISPR/Cas12 can be used to target T-rich regions of the genome which is difficult with CRISPR/Cas9 [44].
Base-editors are another type of genome-editing tools. They are derived from the CRISPR/Cas9 approach and allow precise nucleotide substitutions without double stranded breaks [45]. In plants, DSBs are generally repaired using the NHEJ mechanism which creates small insertions or deletions in the target sequence and usually results in gene knock-out. The alternative HDR mechanism can precisely introduce point mutations using template DNA, however it is very inefficient, especially in plants [45]. Base editors don’t use DSBs, but create precise base substitutions of A-to-G or C-to-T using a nickase Cas9 (nCas) coupled to either an adenine deaminase (ABE) or a cytidine deaminase (CBE), respectively [45, 46]. The CBE-nCa9s fusion converts cytosine to uracil without cutting the DNA and this uracil is later converted to thymine through DNA replication or repair. Similarly, the ABE-nCa9s fusion converts adenine to inosine which is later converted to guanine. A uracil DNA glycosylase inhibitor protein (UGI) can be added to the construct to prevent uracil excision which lowers the efficiency of the transformation [46]. Malabarba et al. [45] applied a CBE base editing system including a UGI sequence and the nCas9-PmCDA1 fusion for the first time in apple and pear on two targets, acetolactate synthase (ALS) and phytoene desaturase (PDS). The authors induced a stop-codon in PDS and an amino-acid substitution in ALS in apple and pear resulting in chlorsulfuron herbicide resistant, dwarfed, albino plants. The study proved the feasibility of targeting multiple genes with base editing in apple and pear but also revealed important challenges that still need to be addressed.
2.4 Limitations of Genome Editing
Despite the advantages and many potential applications of these new genome-editing tools, important challenges still remain. While most studies currently use CRISPR-based methods for genome editing, many of the limitations listed here also apply to ZFN and TALEN methods.
A first major drawback of these genome-editing tools, including CRISPR-based methods, is insufficient target specificity and accuracy. Off-target cleavage can occur when the first 17–20 nucleotides of the sgRNA match with other regions in the genome instead of solely with the target and also in apple, these off-target mutations have been reported [45].
Another problem is the production of chimeras. Several of the studies on the genome editing of pear and apple discussed in this chapter report the production of chimeras during regeneration of transformed apple or pear plants [45]. Chimeras consist of genotypically distinct cells or tissues which may refer to transformed vs non-transformed cells, but also to transformed cells with different mutant alleles where the genome editing machinery has introduced distinct mutations in the same gene of different cells [45]. For example, Charrier et al. [38] reported a high rate of phenotypic chimeras (64% of regenerated transgenic plants) and editing chimeras (88% of pure albino phenotype transgenic plants) after CRISPR/Cas9 KO of the PDS gene in apple. The number of editing chimeras also seemed to increase during the regeneration period, possibly due to the continued expression of the CRISPR/Cas9 cassette. Elimination of chimeras is necessary to establish stable mutants that can reliably pass on the desired trait to progeny. However, exclusion of chimerism through sexual reproduction of the transgenic plants is difficult in the highly heterozygous and self-incompatible woody fruit tree species. Alternatively, adventitious shoot regeneration may be used, but this process takes several rounds of regeneration [45]. Another possible method is the direct delivery of CRISPR/Cas9 ribonucleoprotein (RNPs) into protoplasts which could decrease the number of produced chimera [47]. Direct transfection of apple and grapevine protoplasts has already been performed by delivery of cas9 and gRNA using PEG solution [48, 49].
Transformation efficiency in fruit trees is generally still very low compared to herbaceous plants even using CRISPR/Cas9. One major obstacle contributing to this low efficiency is the highly recalcitrant nature of these species to both genetic transformation and in vitro regeneration [50]. In addition, this transformation and regeneration efficiency seems to vary significantly based on the target, transformed cell type, delivery method, regeneration method, species and even genotype. For example, pear transformation seems to happen at lower efficiency compared to apple: the use of the CRISPR-Cas9 system to knock out Terminal Flower 1 (TFL1) genes in apple and pear to obtain the early-flowering genotype, resulted in 93% of apple transgenic lines compared to 9% of pear transgenic lines [38]. A recent study attempted to increase transformation efficiency in apple through ectopic expression of MdBBM1 which promotes plant regeneration [50]. However, it seems that extensive protocol optimization remains necessary for transformation and regeneration of different species and even different cultivars. This also makes obtaining a transformed plant, albeit less laborious than conventional transgenesis, still very cumbersome, especially since regenerated plants still require screening and quality control.
Finally, there is considerable public concern regarding transgenic plants. After the custom-designed nucleases are transformed into transgenic plants and have induced the desired DSB and mutation, they have to be removed to obtain transgene-free plants. This can happen via genetic segregation and back-crossing, but this method is less suitable for fruit trees [13]. Better approaches include transient expression of CRISPR/Cas9 or the use of preassembled CRISPR/Cas9 ribonucleoproteins (RNPs) [37, 51].
3 Application Potential of Genome-Editing for the Advancement of Important Pome Fruit Breeding Goals
3.1 Yield Improvement
Higher yield is one of the most important traits to achieve in plant breeding, albeit also one of the most difficult since it is a quantitative trait that is determined by many underlying small-effect genes and is highly influenced by environmental conditions and management practices [52]. The complex background of yield makes this trait in itself very difficult to use as a specific breeding target, especially for targeted gene-editing. Instead, it can be considered as a combination of two traits, fruit number (crop load) and fruit size, which need to be carefully balanced and which are both still the result of a complex, multigenic regulation with significant environmental impact.
No known pome fruit breeding programs have increased crop load as a breeding target. Instead, a lot of research is focused on crop load management to minimize biennial bearing and low fruit size due to overcropping. Good management practices to optimize fruit load include cultivar-adapted pruning to optimize the light penetration into the canopy and the distribution of spurs versus extension shoots, thinning of flower buds, flowers and fruitlets, and well-considered root-stock choices to control vegetative growth [53, 54]. High and stable crop load in apple and pear are therefore mainly indirectly targeted in breeding programs through more specific objectives such as root stock breeding, optimized tree architecture and decreased biennial bearing tendency. Tree architecture and biennial bearing are discussed more elaborately later in this chapter.
Similarly, fruit size is also largely determined by environmental factors including orchard management, flower fertilization success, seed number and current crop load [55]. However, more information is available on the genetic basis of fruit size potential compared to crop load since studies often use single fruit weight as an indicative measurement. Fruit size is a complex quantitative trait that is controlled by multiple genes [56]. It depends on the number of cells, the size of the cells and the size of the intracellular spaces [57]. Many QTLs and several major-effect genes have been identified in different studies [56]. Some interesting major-effect genes in apple were homologous to Arabidopsis cell expansion gene AtSAUR19 and tomato fruit size/shape determining genes SlOVATE and SUN [58]. Also, several miRNAs have been associated with fruit weight. For example, overexpression of miR172p in transgenic “Royal Gala” apple significantly reduced fruit size [56]. These miRNAs are part of a large superfamily of transcription factors that play important roles in growth, development and stress response in higher plants [59]. However, the exact functions of most of these genes and miRNAs remain elusive and more research is necessary before they can be reliably used to improve fruit size using genome-editing.
Still, yield as such is often not a primary breeding objective since many other traits are considered at least as important as high yield for the acceptance of a cultivar for commercial production. These traits include many practical aspects of fruit production such as efficient orchard management (trees must be easy to harvest and cheap to maintain), disease resistance, production stability, storability, and transportability. Also many fruit quality characteristics such as flavor, texture, firmness, fruit size and novelty are important for marketability. These traits are breeding objectives in their own right and the most important ones are further discussed in this chapter.
3.2 Fruit Quality Attributes
3.2.1 Sensorial Fruit Quality
The sensory or organoleptic quality of apple and other fresh fruits is determined by three main parameters, namely taste, texture and aroma. These three major sensory fruit quality attributes each independently impact the sensorial evaluation of the fruit, but also exhibit an intricate interplay that largely determines the overall taste and appreciation of fresh fruit. Some organoleptic quality attributes, like soluble sugar content, titratable acid (TA) and fruit firmness can be quantified using biochemical assays. However, these singular quality attributes only provide partial insights of the fruit quality and the evaluation of organoleptic fruit quality is therefore still mainly performed via sensory panels. Research on existing cultivars suggests that, in general, consumers prefer apples with firm, crisp texture, that are moderately juicy and that have a balance of sweet to acid taste. These sensory fruit quality attributes, together with a specific aroma profile, present one of the main targets for consumer-focused plant breeding in apples.
Genetic mapping and association studies in apple and other fruit species have revealed that many of these sensory fruit quality attributes including juiciness, crispness, mealiness, skin color, russet frequency, titratable acidity and soluble solids content have a complex regulation, and are quantitatively determined by a broad range of genomic regions with significant environmental impact [60,61,62,63,64]. This complex regulation with involvement of many genes as well as significant impact from the environment makes it more difficult to improve these fruit quality attributes through targeted gene editing, since (i) key regulatory genes have to be known and sequence-identified, (ii) desired alleles have to be characterized (additivity) together as well as the most optimal combination of alleles at different loci (dominant and epistatic effects), and (iii) all desired alleles have to be genetically engineered within the same generation.
Despite this complex regulation of various aspects of sensorial fruit quality, some major causative genes have been found to regulate specific aspects of fruit taste, texture or aroma and often the most desired allele, i.e. conferring an optimal level of a specific fruit quality parameter, is already known (as well as the genetic source). However, for most of these genes, a specific allelic DNA sequence variant conferring fine-tuned activity of the encoded protein, is desired, rather than a functional knock-out or null mutant, implying that desired traits can only be genetically introduced by advanced editing methods that either introduce nucleotide base pair switches (base editing) or enable allele replacement (HDR – homology directed recombination). These advanced gene editing techniques are only recently available for apple and other perennial fruit trees, and their use and applicability to modulate organoleptic fruit quality has, up till now, not yet been demonstrated. However, studies in other fruit species have shown that targeted editing of specific genes can be used to improve the sensorial quality of fresh fruit. In the following sections, key genes that determine the expression of different fruit quality attributes are outlined together with their desired genetic configuration (allelic variants), with a predominant focus on genes that can be functionally depleted to obtain the desired phenotype.
Fruit taste refers to the basic sensory evaluation of the fruit commodity (sweet, sour, bitter, etc.) and for apple and other pome fruit species mainly depends on the content and composition of soluble sugars and organic acids [65]. Although the overall content of both these types of macromolecules is important, it is mainly the relative sugar/acidity ratio (TSS/acidity balance) of the fruit that determines the fruit’s characteristic taste, implying that both aspects are linked and always need to be improved in parallel. The sweetness of the apple fruit is predominantly determined by sorbitol and the total content of soluble sugars (SSC), with no direct effect of single sugars (like sucrose, glucose, fructose, xylose), though with significant contribution from several volatile compounds, like esters and farnesene [66]. In line with the complex metabolic pathways of each of these sweetness-contributing chemicals, genetic assays have revealed that the sweetness of pome fruit is determined by multiple genetic loci, with each loci only having minor effects [56, 67,68,69,70]. For the fraction of fructose and sucrose in the total sugar pool, however, a major locus was identified on linkage group 1 that respectively explains 47% and 27% of the total variance [67]. This locus harbors the VIN1 vacuolar invertase (MDP0000149570), which enzymatically confers hydrolysis of sucrose into glucose and fructose to regulate the entry of sugars into different metabolic pathways [71]. Similarly, a pedigree-based QTL mapping approach in a “Honeycrisp”-derived germplasm identified and validated three large effect QTLs, i.e. on LG1, 13 and 16, that were consistent across multiple years for the total SSC content in apple fruit [70]. However, the causative genes have not yet been identified. Targeted gene expression studies throughout apple fruit development have shown that the accumulation of fructose in later stages coincides with an enhanced expression of the tonoplast monosaccharide transporters (TMTs) MdTMT1 and MdTMT2 which convert the excess amount of imported sugars into starch. At final fruit maturation, the accumulation of sucrose overlaps with an elevated expression and enhanced activity of the sucrose-phosphate synthases (SPS) MdSPS5 and MdSPS6, which catalyze the transfer of a hexosyl group from UDP-glucose to D-fructose 6-phosphate to form UDP and D-sucrose-6-phosphate [72]. A combined QTL and transcriptomics study in Asian pear (Pyrus pyrifolia) identified two sucrose transport genes (PpSUT, LOC103964096, and LOC103940043) that are negatively correlated with the sugar content in ripening fruit [73]. In addition, two sorbitol dehydrogenase genes (PpSDH genes, LOC103960512 and LOC103960513), were also found to be negatively co-expressed with total sugar content in the fruit, indicating that these act as antagonists of fruit sweetness [73]. Plants harbor different types of sugar transporters, and the SWEET-class of sugar transporters (i.e., Sugar Will Eventually Be Exported Transporters) have been found to play a major role in sugar accumulation in the fruit of apple as well as various other fruit tree crops. Marker-based association studies thereby revealed that in particular three SWEET genes, i.e. namely MdSWEET2e, MdSWEET9b, MdSWEET15a, are significantly associated with total sugar content in the fruit, with MdSWEET15a and MdSWEET9b accounting for a relatively large portion of the phenotypic variation (and located on a region harboring a QTL for sugar content) [74, 75].
Fruit acidity is a major determinant of the overall apple flavor and strongly influences the perception of other flavor traits such as sweetness and aroma. The pH of fresh apples generally ranges from 3.3 and 4.0, making them mildly acidic, and this is almost exclusively attributed to the accumulation of organic acids, with malic acid forming the major fraction (±90% of all organic acids) followed by quinic acid (±5%), citric acid (±1,5%) and small amounts of ascorbic, shikimic, maleic and tartaric acid [76, 77]. While in wild apple varieties fruit acidity is determined by the content of both malic and citric acid, in cultivated apple varieties citric acid is almost completely absent and fruit acidity is almost exclusively determined by malic acid [76, 78]. The concentration of malic acid can be measured sensorially through panel tasting, or more quantitatively by pH measurement, analytical methods (HPLC) or via titration of fruit juice, with the latter actually indicating the total content of organic acids as expressed in malic acid equivalents per fruit mass (TA: titratable acidity) [79]. Although acidity of freshly harvested apples is a complex trait, genetic studies have revealed that it is predominantly determined by two large effect QTLs together with various minor QTLs that mainly determine the variance in high acidity apple varieties (e.g. Ma4, Ma6, M2, and M3) [80,81,82,83]. More specifically, apple fruit acidity is genetically determined by one QTL on LG16, referred to as the Ma locus [84], and one on LG8, called the Ma3 locus [85], with both QTLs jointly explaining 66% ± 5% of the phenotypic variation through an additive allele dosage model with incomplete dominance [86,87,88]. For the Ma3 locus on LG8, up till now, no candidate genes have been identified that control apple fruit acidity. In contrast, high resolution mapping and expression assays provided strong evidence that the aluminum-activated malate transporter gene (ALMT1 or Ma1) is the fruit acidity determining-gene in the Ma locus [89]. ALMT1 encodes a membrane-associated protein that is targeted to the tonoplast and actively transports malic acid molecules from the cytosol to the vacuole, which serves as major subcellular repository for organic acids, hence contributing to its cellular accumulation [90]. The low acidity ma1 allele still localizes to the tonoplast but exhibits reduced malate transport functionality as compared to the pseudo-dominant high acidity Ma1 allele due to a 84-AA truncation in the conserved C-terminal end domain [90]. Diversity studies in apple also showed that expression of Ma1 is significantly correlated with the fruit titratable acidity at harvest [89], making it a highly suitable candidate for direct modulation of fruit acidity through gene editing approaches. Recent studies provided more insights into the regulation of these tonoplast transporters in apple fruit and identified several MYB transcription factors, including MdMYB1 and MdMYB73, as important regulators of vacuolar accumulation of malate. MdMYB1 promotes the expression of two genes encoding B subunits of vacuolar H+-ATPase (VHA), MdVHA-B1 and MdVHA-B2, and thereby transcriptionally activates its H+ pumping activity and enhances the transport of malate into the vacuoles [91]. Similarly, MdMYB73 transcriptionally activates MdALMT9, MdVHA-A and MdVHP1 (vacuolar pyrophosphatase 1) to enhance their activity, leading to increased concentrations of malate and vacuolar pH [92]. The activity of MdMYB73 towards its downstream targets was thereby found to be positively influenced by the interaction with MdCIbHLH1. This transcriptional cascade (that promotes malate accumulation in the vacuole) is antagonized by the BTB-BACK-TAZ domain protein MdBT2 which targets MdCIbHLH1 and MdMYB73 for ubiquitination and subsequent degradation by the 26S proteasome pathway, hence negatively regulating accumulation of malate and vacuolar acidification [93]. Recently, another R2R3 − MYB transcription factor, namely MdMYB44, was found to control the fruit malate content and acidity in apple, though in a negative manner. MdMYB44 represses the promoter activity of the malate-associated genes Ma1 (Aluminum-Activated Malate Transporter 9), Ma10 (P-type ATPase 10), MdVHA-A3 (V-type ATPase A3), and MdVHA-D2 (V-type ATPase D2), and thus suppresses vacuolar import and accumulation of malate. Importantly, specific SNPs in the promotor region of MdMYB44 thereby showed strong association with fruit malate content, i.e. either through their effect on basal activity or by altering affinity towards the basic-helix–loop–helix TF MdbHLH49 [94]. Parallel to MdMYB44, the protein phosphatase MdPP2CH also negatively regulates accumulation of malate in the fruit by post-transcriptionally suppressing the activity of the vacuolar H+-ATPases MdVHA-A3, MdVHA-B2 and MdVHA-D2 as well as the malate transporter MdALMTII through dephosphorylation [95]. As MdSAUR37 was thereby found to promote malate accumulation in the apple fruit by negatively regulating the MdPP2HC phosphatase activity, the MdSAUR37/MdPP2CH/MdALMTII chain was found to precisely determine apple fruit malate contents through hierarchical epistatic genetic effects [95]. Overall, multiple genetic factors that contribute to metabolism and vacuolar accumulation of organic acids, and particularly malic acid, in apple fruit have been retrieved with identification of both positive and negative regulators and associated characterization of allelic effects. Despite the absence of concrete examples, these genetic insights provide a strong basis for precisely modulating the titratable acidity as well as the overall organoleptic appreciation of the apple fruit through targeted gene editing approaches such as CRISPR and TALENs.
3.2.2 Nutritional Quality and Food Functionality
During the last decade there has been a paradigm shift regarding consumer acceptance towards fruits. Due to the general awareness of the impact of food consumption on personal health and overall increased welfare, consumers now also take into account the nutritional, functional, and physio-chemical factors of fruits, in particular for fresh produce. Also for pome fruit, such as apple and pear, there is an increasing preference for varieties that have high levels of health-promoting compounds, such as essential vitamins, minerals, dietary fibers, antioxidants and other key phytochemicals. The visual characteristics (e.g. skin characteristics), eating quality (e.g. texture and flavour), and storability are among the main fruit quality traits being targeted in apple breeding programs, but the enhancement of phytochemicals is now gaining traction to select “bio-fortified” apple cultivars. However, despite their relevance for public health, almost no directed selection for specific fruit bio-chemicals or overall augmented nutritional value has yet been performed. As a result, current apple varieties generally do not have high food functionality, and instead, for most nutritional parameters, have reduced contents or values as compared to their wild counterparts. For example, several studies reported that modern apple varieties have drastically reduced polyphenol content (particularly stilbenes, hydroxycinnamic acids, and dihydrochalcones) compared with the ancestral heritage, wild progenitors (Malus sieversii) and germline cultivars [96, 97]. Similar observations were made for organic acids, including malic and ascorbic acid, indicating significant counter-selection during domestication and breeding, most likely as indirect effect of selection against bitterness [96]. In contrast, some relevant flavonoids (flavonols and flavan-3-ols) and triterpenoids (ursolic, oleanolic, and betulinic acids) did not show this selection-induced reduction in modern apple varieties [96]. Moreover, the few incentives that projected to increase the nutritional fruit quality in apple via conventional ways failed or only had limited success, mainly due to the complexity of the underlying biochemical pathways and adverse side effects on other agronomic or consumer-related attributes.
Now, gene editing approaches allow specific enhancement of the health-related nutritional composition and food functionality of commercial apple cultivars, without affecting their typical flavor and fruit quality attributes, and therefore form an easy and straightforward method to improve general public health. Germplasm characterization studies have revealed dramatic variation in the content of various nutritional compounds in apple, including polyphenols, vitamin C, etc., indicating that the biochemical composition of the fruit is predominantly genetically determined. However, genetic studies have shown that both the content and composition of these phytochemicals is generally under polygenetic control with multiple small effect genetic loci. This diluted genetic control largely impairs the genetic enhancement of these compounds through single gene editing approaches. Though, for some biochemicals, like polyphenols, the polygenetic control is mainly determined by a small number of genetic loci that have a small effect [98]. In these cases, knowledge of these genes (the relatively simple genetic architecture) may provide a basis to significantly optimize the content or composition of the specific compound of interest through targeted gene editing approaches. For many fruit bio-chemicals, underlying genetic loci (QTLs) and associated candidate genes have been identified in apple through genetic linkage mapping or association studies [63, 99, 100]. Using a combined genomics-metabolomics approach, both Khan et al. [100] and Bilbrey et al. [99] hereby detected a large number of metabolite quantitative trait loci (mQTL) spread along all chromosomes, with hot spots on the linkage groups 16 and 17 for apple phytochemicals. However, up till now, only a few regulatory genes have actually been identified and were validated to play a functional role in the determination of the biochemical composition of apple fruit.
For polyphenols, candidate genes for the production of quercetin, epicatechin, catechin, chlorogenic acid, 4-O-caffeoylquinic acid and procyanidins B1, B2, and C1 have been retrieved via mapping [98], however, no further functional validation has yet been performed and actual genetic regulators have not yet been identified. Specifically for the dihydrochalcone phloridzin (phloretin 2′-O-glucoside), i.e. the most abundant phenolic compound in apple trees (Malus × domestica), regulatory enzymes have been identified via genomics and in vitro studies with validation through transgenic approaches. In particular, six glucosyltransferases (UGTs) have been identified which are able to selectively glucosylate phloretin, i.e. the direct precursor of phloridzin [101]. As a follow up, one recent study used genetic approaches, including RNAi and CRISPR, to analyze the function of one of these UDP-2′-O-glucosyltransferases, namely MdPGT1, in phenol metabolism, and thereby demonstrated that PGT1 stimulates the production of phloridzin in the leaves of apple with distinct morphological differences between knock-down and genome-edited mutant lines [102].
Anthocyanins form an important group of phenolic compounds, as they confer health-related benefits due to their antioxidant activity but also contribute to sensorial fruit acceptance due to their role as a pigment. Several molecular regulators of anthocyanin metabolism have been identified, though most act in the biosynthesis and thus have a promotive effect on anthocyanin accumulation. For example, the anthocyanin biosynthesis pathway is transcriptionally regulated by the MYB-bHLH-WD40 (MBW) complex, with allelic variants of the enclosed MYB10/MdMYB1 TF [103] determining tissue-specific expression and thus controlling apple fruit (peel and flesh) as well as foliage color. Overexpression of MYB10 leads to a significant increase in foliar, flower and fruit anthocyanins, especially in the fruit peel, with no negative impact on sensorial quality and other consumer-related quality traits. However, due to their promotive effect, these regulatory genes do not form suitable candidates for CRISPR-based gene editing for enhancing anthocyanin contents. Besides these promotive proteins, two other MYBs have recently been identified as transcriptional inhibitors of anthocyanin biosynthesis; namely MdMYB6 and MdMYB306. Xu et al. (2020) showed that MdMYB6 inhibits anthocyanin synthesis by directly inhibiting MdANS and MdGSTF12, i.e. two positive regulators of anthocyanin production, and by reducing contents of the precursors UDP-glucose and UDP-galactose by regulating the monosaccharide transporter MdTMT1 [104]. In addition, a second R2R3-MYB TF, namely MYB306-like, was found to act as an anthocyanin repressor gene. More specifically, the MdMYB306-like protein activates the expression of an anthocyanin repressor gene, MdMYB17, and inhibits the expression of the anthocyanin structural gene MdDFR through direct promotor binding, and additionally interacts with MdbHLH33 and MdMYB17 to enhance its TF regulatory activities [105]. In line with this, transient silencing of MdMYB6, MdBY306-like and MdMYB17 leads to increased anthocyanin concentrations, indicating that these genes form interesting targets for CRISPR-based mutagenesis to obtain increased anthocyanin contents in apple fruit [105]. Besides these antagonistic regulators of anthocyanin identified in apple, genetic studies in various other fruit crop systems, i.e. in particular tomato, have also identified several other factors that operate in the flavonoid metabolic pathways to suppress or reduce anthocyanin production. These include the ATROVIOLACEA (ATV) R3-MYB protein [106] and the nuclear protein DE-ETIOLATED1 (DET1) [107]. Genetic loss of function of these proteins is associated with a significant increase in anthocyanins in the mature fruit, making them excellent candidates for CRISPR based mutagenesis of their orthologous proteins in pome fruit for increasing anthocyanin contents in the peel and the pulp.
Another bio-chemical component with strong nutritional value as antioxidant is vitamin C or ascorbic acid (AsA). Besides its general role in oxidative stress mitigation, dietary L-AsA also has various other important health benefits. Increased intake of vitamin C has been associated with a decreased incidence of several important human diseases, such as cataract, cardiovascular diseases, and cancers. Vitamin C also promotes the uptake of iron and zinc, which is particularly relevant in meat-poor diets. Humans cannot synthesize Vitamin C due to the absence of the gene encoding L-guluronic acid-1,4-lactone oxidase, which catalyzes the last step in the AsA synthesis pathway, and therefore completely rely on dietary intake of AsA to meet their daily requirements [108]. Fruits are the main source of human AsA intake, though fruit AsA levels in commercial pome fruit cultivars are generally quite low as compared to other fruit species, such as lemon, orange and strawberry, with apple and pear only containing 0.05–1.0 and 5–10 mg AsA per 100 g fresh weight, respectively [109]. The ascorbic acid metabolic pathway has been extensively studied in plants, and has been found to be rather complex involving several parallel pathways that include multiple enzymatic steps [108], with total AsA accumulation being regulated by transcription factors, protein interactions, phytohormones, and environmental factors. Despite the fact that most of the regulators have already been identified in model systems, their functional role and contribution in the AsA metabolism of pome fruit has not yet been resolved, although there are a few exceptions. For example paralogs of GDP-l-Galactose Phosphorylase (GGP) have been found to act as a major determinant of Vitamin C concentration in apple fruit, with specific alleles leading to a significantly higher level of Vitamin C content in the pulp [110]. However, as these alleles do not confer a functional GGP knock-out, but instead promote AsA biosynthesis through formation of specific protein variants, these GGP paralogs do not form suitable candidate genes to boost AsA levels in the pome fruit via CRISPR mutagenesis. Therefore, it is still unclear which genetic factors can be used for the engineering of increased fruit AsA levels in pome fruit through CRISPR editing.
3.3 Agronomic Traits
3.3.1 Disease and Pest Resistance
Apple and pear orchards are routinely plagued by insect pests, including aphids, mites, and caterpillars. Also many bacterial and fungal diseases are prominent in pome fruits. By far the most destructive bacterial disease is fire blight (Erwinia amylovora). Major fungal diseases include apple scab (Venturia inaequalis), pear scab (Venturia pirina and Venturia nashicola) and powdery mildew (Podosphaera leucotricha). Viral diseases can generally be sufficiently controlled through the use of certified virus-free plant material. Economic losses due to these and other pests and diseases include loss of produce, reduced fruit quality, loss of trees or even orchards, disruption of orchard production, management costs, and costs related to stringent quarantine and international trade regulations. In addition, preventive and curative treatments through chemical fungal sprays or antibiotic sprays raise important environmental, biosafety and health concerns [111,112,113,114]. Disease and pest resistance is therefore an important breeding objective in pome fruit species.
A first approach to improve disease resistance is through the introgression of resistance genes into elite cultivars. There are two main types of disease resistance, namely quantitative and qualitative resistance. Quantitative resistance is conferred by many small-effect genes and results in partial resistance to multiple pathogen strains making it highly durable [111]. On the other hand, qualitative resistance is usually based on a gene-for-gene interaction between the pathogen avirulence (Avr) gene and the plant resistance (R) gene and often leads to a hypersensitive response against the pathogen. This form of single major gene disease resistance is easier to obtain for breeders, but can also be more easily broken by newly evolving virulent pathogen strains [111, 115]. It is therefore preferred to stack disease resistance genes or to combine qualitative and quantitative resistance [111, 114]. Resistance genes have been identified for several important pear and apple diseases. For fire blight, the only functionally characterized resistance gene is FB_MR5, however resistance conferred by this gene has regrettably already been overcome [116, 117]. Several other putative candidate resistance genes include FB_Mfu10, NBS-LRR genes of ornamental cultivar Evereste, and several genes with disease-related domains in the FB_Mar12 region [117,118,119]. For apple scab resistance, around 20 R genes are known, but not all confer equally durable resistance. Through worldwide monitoring of Rvi breakdown, the genes Rvi5, Rvi11, Rvi12, Rvi14 and Rvi15 were identified as rarely overcome, possibly because of an associated fitness cost to the pathogen [120]. In pear, identified resistance genes for Venturia nashicola include RVnk [121], Rvn2 [122] and Rvn3 [123]. For V. pirina, Rvp1 was identified [124]. Powdery mildew is mainly studied in apple where several resistance genes are known: Pl1, Pl2, Plw, Pld [125]. However, current advanced breeding material generally lacks major resistance genes and therefore, wild Malus and Pyrus species remain the most important source of disease resistance alleles. This hampers application of conventional breeding because of problems with the long life cycle of pome fruit species and linkage drag [111], especially when pyramiding multiple resistance genes. This process can be sped up through transgenic approaches. There are several examples of genetic transformation to bring resistance genes into apple and pear [126,127,128]. However, this approach is more difficult to apply using gene-editing techniques which are more suited for knock-out of negative regulators of resistance.
Alternatively, disease resistance in plants may be achieved through the silencing or knock-out of susceptibility (S) genes. Several studies have successfully obtained resistance cultivars through genetic transformation with S genes as target. For example silencing of the HIPM gene (HrpN-interacting protein from Malus) through RNA interference resulted in increased fire blight resistance in apple cv. “Galaxy” [129]. Knockout of susceptibility genes is well suited for genome editing applications and has already been applied in apple cultivars “Gala” and “Golden Delicious” to significantly reduce bacterial fire blight symptoms through inactivation of MdDIPM4 which interacts with pathogen effector protein DspA/E [37, 130]. Similarly, another study targeted DIPM1, DIPM2 and DIPM4 in “Golden Delicious” to increase resistance to fire blight [49]. Susceptibility genes don’t always have to encode proteins that directly interact with plant effectors, but may also be more broadly involved in plant immunity. For example, targeted silencing of a frequently used susceptibility gene MdMLO increased powdery mildew resistance in “Gala” [131]. MLO is a transmembrane protein located in the plasma membrane and is presumed to be involved in plant defense and immunity responses [132].
Plant susceptibility genes may also be found for insect pests. For example, such genes may be involved in the induction of defense signaling pathways, food accessibility and food quality [133]. For example, in A. thaliana, the transcription factor WRKY22 is involved in pathogen-triggered immunity and knock-out mutants were more difficult to colonize by aphid populations [134]. However, knowledge of S-genes against insect pests is limited and to our knowledge there have been no applications in pome fruit species to date.
Finally, another alternative approach is to target pathogen or microbial genomes using CRISPR/Cas9 technologies as compared to focusing solely on the host. For example, the gene drive system, which was shown to be successful in mosquito [135] may be adapted and applied in sexually inheriting plant pathogens [136]. Original gene drive systems are based on homing endonuclease genes (HEGs) which encode proteins that cleave a recognition site of around 20–30 nucleotides on the genome. The HEG itself is inserted in the middle of its own recognition site thereby protecting itself from further cleavage. When the HEG comes in contact with the target site on the wild-type homologous chromosome it will induce a DNA DSB. The DSB is repaired using the homologous chromosome as a template and the HEG allele is copied into the broken chromosome. As a result, these HEGs can spread rapidly through populations [137]. The gene drive system can be made more accurate and flexible using CRISPR-based adaptations [138] and there are many agricultural applications, including the sensitization of a population to pesticides (local sensitizing gene drive), manipulating plant pathogen-vector relationships, and knock-out of virulence genes [139]. The latter has already been shown successful under in vitro conditions for the wheat pathogen Fusarium graminearum [140]. Similarly, the CRISPR/Cas9 gene drive approach may also be applied to enhance performance of biocontrol agents or beneficial organisms which can heighten plant immunity [136]. However, this system is subject to major concerns regarding biosafety and bioethics and should not be applied without extensive risk assessment [141].
3.3.2 Abiotic Stress Tolerance
As sessile organisms, plants continuously endure a whole range of environmental stressors including drought, waterlogging, extreme temperatures, soil salinity, wind, and hail, which can severely affect growth and production. Improved abiotic stress tolerance allows fruit trees to be grown in suboptimal environments which is especially relevant in light of climate change and the associated occurrence of more extreme weather events worldwide. Important breeding objectives include tolerance to drought, cold, and soil salinity.
Drought tolerance in apple and pear trees is often tackled through root stock breeding in an attempt to achieve enhanced uptake of water through a better-developed root system. A recent study describes the knock-out of GRETCHEN HAGEN3.6 (MdGH3.6), an indole-3-acetic acid conjugating enzyme, in apple rootstocks through RNAi to increase tolerance to prolonged drought periods without impacting fruit quality. Knocking out this gene increased IAA content, adventitious root number and root length [142, 143]. Other potential drought tolerance related targets for CRISPR genome editing that were successful in other species include genes involved in stomatal density (VvEPFL9-1) [144], stomatal response (AtOST2) [145, 146], genes involved in ABA signaling (AtAREB1, SlMAPK3, OsSAPK2), and leaf rolling (OsSRL1 and OsSRL2) [145].
Although adult pome fruit trees are generally quite cold-hardy, severe production losses can still occur due to frost damage to flower buds, flowers, young shoots and fruits. Cold hardiness can differ between cultivars [147], but little is known about the genetic and biochemical basis. Several genes for general cold-hardiness have been identified in apple. Putative positive regulators of cold hardiness in apple include MdHYL1, Mdm-miR172 [148], and CBF genes (C-repeat binding factors) like MdCBF1-5 [149]. Knock-out of negative regulators, such as Mdm-miR156, can cause increased cold-hardiness, which is more interesting for genome editing application [148]. Many of the genes and pathways involved in cold-tolerance are also involved in other (a)biotic stress responses. For example, the previously mentioned CBF genes are considered hub genes that are involved in drought, salinity and cold responses [149, 150]. Targeting these genes may result in simultaneously increased tolerance to other abiotic stress factors, however caution must be taken to avoid undesired side effects.
Heat stress in pome fruits can lead to sunburn of leaves and fruits, scorching of leaves and in extreme cases early fruit drop and even leaf drop [151, 152]. Damage can be especially severe in combination with drought. In high temperature conditions, damage may occur in plant cells due to protein misfolding and denaturation, damage to membranes and accumulation of ROS species [153]. Gene targets to enhance heat tolerance in pome fruits are likely to be found in plant antioxidant defense systems or osmotic adjustment pathways [154]. Heat stressed plants will also generally synthesize a variety of heat-shock proteins (HSP). Such HSPs are also found in apple and have been identified as important regulators of temperature stress responses. Members of the HSP20 gene family appear to be especially important and are highly upregulated in response to heat stress [151]. Another heat-stress related gene is MdPRP6 which was shown to enhance heat stress tolerance when overexpressed in transgenic tobacco plants [153]. Possible genome editing targets are negative regulators of these heat shock related genes.
Finally, soil salinity is detrimental to plants due to osmotic stress and salt toxicity [155]. It mainly occurs due to irresponsible irrigation and fertilizer application. The plant may try to overcome salt stress by inducing osmoregulating and antioxidant systems [156]. In apple, MdINT1 has been shown to confer salinity tolerance by regulating antioxidant systems, and homeostasis of ions and osmosis [156]. Several other genes involved in response to salt stress have been identified in other plants, but relatively little knowledge is available in apple and pear. For example, knockout of AITR (ABA induced transcription repressors) gene family members in A. thaliana increased drought and salinity resistance [157]. Also in A. thaliana, a nucleoporin gene NUP85 seems to modulate the response to salt stress [158]. Improved salt tolerance was also achieved in rice by CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene involved in cytokinin transduction and metabolism [159], and in tomato through down regulation of Auxin Response factor 4 (ARF4) [160]. Homologous gene targets may be explored in pome fruit species.
3.3.3 Tree Architecture
Tree architecture is influenced by four main factors: primary growth, branching patterns, flowering location and meristem and shoot mortality [161]. In commercial pome fruit production, tree architecture can influence important factors such as fruit quality, yield and orchard management requirements including planting density, pesticide application efficiency, harvesting efficiency, and requirements for thinning, pruning, branch-bending and tying [161]. The ideal tree form would allow high density plantings and maximum automatization [162]. The main breeding goals regarding tree architecture in pear and apple are dwarfing (mainly achieved through dwarfing root stocks) and optimal branch orientation [163]. Causative genes have been identified for the dwarfing trait, branch orientation and the columnar growth habit in apple [162].
Dwarfing root stocks limit tree size, enable high density plantings, increase flower density and allow more efficient mechanization [164]. Mainly genes associated with growth-related plant hormones are linked to the dwarfism trait and may be identified as putative targets of gene editing. For example, in peach, a nonsense mutation in the GA receptor GID1c was found to result in dwarfism [165]. In apple, overexpression of the transcription factors WRKY9 and NAC1, which are negatively involved in the brassinosteroid biosynthesis pathway, was shown to result in dwarfing [162, 164, 166].
Several gene families are associated with branch orientation, namely WEEP, and IGT family genes TAC1 and LAZY1 [167,168,169]. Tiller Angle Control1 (TAC1) promotes lateral shoots to grow outward and reduced or eliminated expression of this gene causes more upright growth habits [162]. In contrast, LAZY1 promotes upward shoot orientation. Plants with reduced or no LAZY1 expression show wide shoot angles [170, 171]. A weeping growth habit was found in birch and plum with a defect or silenced LAZY1 gene [162, 172]. In theory, a desired branching phenotype may be obtained in trees by balancing the effects of targeted editing of one or several of these genes.
One of the most ideal branching habits in apple is the columnar type. These trees grow upwards as a column with very short branches and an increased number of spurs [162]. They are ideally suited for high density plantings and automatization, although they are generally very susceptible to biennial bearing and produce lower quality fruits [173,174,175]. Columnar growth habits have been achieved in apple through mutation of the MdCoL gene, which encodes a putative 10G-FEII oxygenase [161, 176], and overexpression of LEAFY [177].
3.4 Pollination and Fertilization
3.4.1 Self-Incompatibility
Generally, fruit set in pome fruit species is dependent on cross-pollination between two cross-compatible cultivars due to the S-RNase dependent gametophytic self-incompatibility system (GSI) which prevents self-fertilization [178,179,180]. This system is genetically controlled by the S-locus which carries a pistil-expressed S-RNase gene and multiple pollen-expressed SFBBs (S-locus F-box brothers) [178, 181,182,183]. According to the currently accepted non-self-recognition mechanism, SFBB proteins expressed by the pollen haplotype recognize non-self S-RNases in the style, but not self-S-RNases, and mediate their degradation through the ubiquitin-26S proteasome [184, 185]. Problems with this innate self-incompatibility arise when cross-pollination is hampered. To ensure sufficient cross-pollination and thus economically viable yields, several conditions must apply including (1) adequate presence of a diverse pollinating insect population, (2) presence of well-distributed trees of a cross-compatible pollen donor cultivar with overlapping flowering period in the orchard, and (3) optimal weather conditions during flowering. However, these conditions cannot always be met and even then, fruit set after self-pollination is expected to be more stable compared to cross-pollination [186]. Therefore, self-compatibility (SC) has become an important breeding objective [5].
Spontaneous self-compatible (SC) mutant apple and pear varieties are rare and hardly ever suitable for commercial fruit production. One exception is “Osanijisseiki” which is a natural SC mutant of the Pyrus pyrifolia cultivar “Nijisseiki”. “Osanijisseiki” was released as cultivar in 1979 and has been used as a parent to breed new SC cultivars using conventional breeding strategies [5]. Spontaneous SC mutants in pome fruit species are either the result of pistil-function breakdown, essentially meaning knock-out of the S-RNase gene [181, 187, 188] or alternatively of competitive interaction due to polyploidy or segmental duplications in the S-locus [189, 190]. The latter approach is less suited as genome-editing application, however pistil-function breakdown of self-incompatibility has already been successfully applied in apple to create a transgenic SC Elstar mutant using a co-suppression approach resulting in S-RNase gene silencing [186]. Knock-out of the S-RNase gene could be performed relatively easily using genome-editing approaches to introduce the self-fertility trait into established commercial cultivars.
3.4.2 Parthenocarpy
Parthenocarpy refers to natural or artificially induced fruit development without fertilization of the ovule [191]. In commercial pome fruit production, this is a very interesting trait because it can alleviate problems with pollination, self-incompatibility, biennial bearing and spring frost. In addition, parthenocarpic fruits are seedless and often have more edible pulp and less core which is often preferred by consumers and can be an advantage for industrial food processing applications [192, 193]. Parthenocarpy is mainly genetically determined and can vary between species and cultivars. In pear, natural parthenocarpy is more common in Pyrus communis cultivars compared to the Asian pear species, such as Pyrus pyrifolia [194]. Pyrus communis parthenocarpic cultivars include “Conference” [195] and “Bartlett” [196]. In apple, some parthenocarpic fruit can develop on “Cox’s Orange Pippin”, “Wellington Bloomless” and “Spencer Seedless” [197].
Natural parthenocarpic fruit set in pome fruit species is usually relatively low, but can be stimulated using plant growth regulators (PGR) during early fruit development to obtain economically viable yields. These PGRs generally impact the auxin, cytokinin or gibberellin pathways that are involved in fruit set and development [193, 198, 199]. Correspondingly, parthenocarpic mutations in a variety of species are generally found in the synthesis and metabolism pathways of these hormones [193]. Overexpression of auxin biosynthesis or receptor genes and silencing of auxin signal repressor genes or negative regulators of auxin signaling are associated with parthenocarpy in tomato (Solanum lycopersicum) and eggplant (Solanum melongena) [193, 200,201,202,203]. In the gibberellin pathway, overexpression of GA biosynthesis genes, such as gibberellin 20-oxidase genes (GA20ox), and suppression of GA respressor genes like DELLA and GA2ox genes have been shown to lead to parthenocarpy in A. thaliana and tomato [204,205,206,207,208].
Alternatively, floral homeotic genes are also often associated with parthenocarpy. For example, the silencing of genes responsible for stamen identity has been associated with parthenocarpy in tomato, possibly because stamens act as negative regulators to restrict ovary development before pollination and fertilization have occurred [209, 210]. These genes include, amongst others, several class B MADS-box genes such as TOMATO APETALA3 (TAP3), DEFICIENS (slDEF), TOMATO MADS BOX GENE6 (TM6), and TOMATO PISTILLATA (TPI) [209, 211,212,213]. In apple, two parthenocarpic cultivars showed similar splicing variants of the AtPI homolog which were causally related to their parthenocarpic trait [214, 215]. In pear, several transcriptomic studies on induced parthenocarpic fruit development in pear showed possible involvement of homologs of many of the above listed tomato genes. For example, key GA-associated genes related to parthenocarpy in pear include GA20ox, GA3ox, GA2ox, GA receptor GID1, and DELLA [216, 217], as well as MADS-box class B gene DEF. However, the genetic basis for parthenocarpic fruit set is still largely unclear in woody fruit tree species and mutations in many of the mentioned genes cause undesired pleiotropic effects. In addition, pathways leading to parthenocarpy in pome fruits may be different compared to A. thaliana which produces siliques or tomato which produces botanical fruits compared to accessory fruits. Therefore necessary caution must be taken when choosing possible gene targets for parthenocarpy in pome fruit trees based on studies in these model species.
3.5 Tree Phenology
As perennial species, pome fruit trees require immaculate regulation of their phenology to survive and reproduce in temperate climates with seasonally changing climatic conditions. In order to anticipate these seasonal changes, temperate tree species take climatic cues to regulate important transitions in their life-cycle, including dormancy, bud burst, flowering, fruit development, and leaf-drop. Phenology characteristics greatly determine the success of a cultivar in a given location due to their impact on the tree life-cycle, including many reproductive traits that are essential for fruit production. As a result, considerable effort has been devoted to elucidate the molecular mechanisms that underly important phenological transitions. In the following paragraphs three aspects of tree phenology are briefly discussed which have major relevance to pome fruit production: juvenility, dormancy and biennial bearing. We also discuss potential applications of gene editing techniques based on current knowledge.
3.5.1 Juvenility
Juvenility in most pome fruit species can take 5–6 years which significantly extends the breeding cycle and delays research on reproductive biology [218]. Therefore, a shortened juvenile period is of great interest to breeders and researchers, and several attempts have been made to obtain early flowering apple and pear mutants with the goal of accelerating conventional pome fruit breeding and research. For example, apple and European pear TFL1-1 mutants showing an early flowering phenotype were successfully obtained using CRISPR-Cas9 technology [38]. These mutants flower continuously in vitro, do not require cold accumulation to induce flowering and completely by-pass the juvenile period. However, the benefits of cultivars with a short juvenile period are limited in the context of fruit production, since mature tissue can be clonally propagated onto rootstocks once juvenility is broken.
3.5.2 Dormancy
To survive the harsh winter conditions, temperate fruit trees go through both endo- and ecodormancy. Dormancy can be defined as the “temporary suspension of visible growth of any plant structure containing a meristem” [219]. In pome fruits, both mixed and vegetative buds are formed during the flowering period [220]. In autumn, low temperatures and short day conditions induce a state of endodormancy in these buds which can only be broken by the fulfillment of the cultivar-dependent chilling requirement [221]. Once this chilling requirement is reached, buds transition into a state of ecodormancy. Ecodormancy is broken by higher temperatures, initiating bud break. This two-factor regulatory system prevents early bud break on a warm day in late autumn or during cold days in early spring [222]. Ideally, this break of two dormancy states initiates uniform flowering, which is advantageous to both the plant and the grower. Dormancy cycles can be disturbed when pome fruit trees cannot reach the required amount of chilling after a mild winter. This can lead to abnormal bud break, resulting in extended flowering periods, delayed leaf formation and asynchronous fruit development [223]. Currently, problems associated with abnormal bud break are mainly observed in orchards grown in subtropical climates, but may also start to occur in more temperate regions as a consequence of climate change. Several chemical treatments can be used to break dormancy in pome fruits including Dormex, potassium nitrate, and mineral oil [224]. However, these products are associated with environmental and health concerns and pose significant costs to growers. Therefore, the development of commercial apple and pear cultivars with low chilling requirements would be beneficial.
Several key hormones and some regulatory genes involved in the maintenance and release of endodormancy have been identified [222]. In pear, abscisic acid (ABA) levels increase during endodormancy induction and remain high during endodormancy. During cold accumulation, transcription of PpCYP707A-3, which encodes an ABA 8-hydrolase enzyme, sharply increases. Simultaneously, ABA levels in the buds decrease initiating transition from endodormancy to ecodormancy in pear [225]. The onset of this transition is believed to result from the release of inhibition on GA biosynthesis, regulated by PpGAST1. Transcription of PpGAST1 is inhibited by ABA and decreasing ABA levels during dormancy transition allow PpGAST1 levels to rise. Increased PpGAST1 transcription levels were accompanied by increased transcription of PpGA20OX2, a GA biosynthesis gene. Additionally, high ABA levels also indirectly induce the GA catabolism gene PpGA2OX1, resulting in decreasing GA levels [226]. Another important gene group associated with dormancy regulation is the dormancy-associated MADS-box (DAM) gene family. Their expression is tightly regulated by ABA through several transcription factors including ABA response element (ABRE)-binding transcription factor 1 (AREB1), which represses PpDAM1 transcription in pear. PpDAM genes promote ABA biosynthesis by upregulating the expression of PpNCED3, an ABA biosynthesis gene [227]. In transgenic pear calli, DAM3 was found to also inhibit cell division and cell growth, supporting their role in pear bud dormancy. Interestingly, two Asian pear cultivars (P. pyrifolia) “Suli” and “Cuiguan” with respectively a high and a low chilling requirement, showed different expression patterns of PpDAM3 during endodormancy [228]. Additionally, epigenetic regulation is also found to be involved in bud dormancy. In peach (Prunus persica) and sweet cherry (Prunus avium), DAM genes were found to be under epigenetic regulation by histone modifications and/or DNA methylations during the dormancy process [222]. In apple and pear, bud break was associated with a decrease in DNA methylation under ideal high chill conditions [229, 230]. However, the precise mechanisms underlying epigenetic regulation of dormancy in pome fruits remain unknown.
As negative regulators of dormancy release, DAM genes appear to be good candidates for targeted gene editing. DAM gene expression may be altered through CRISPR transcriptional repression to adapt the cold requirement of various pome fruit cultivars. Alternatively, CRISPR-Cas9 may be used for targeted mutagenesis of the DAM genes. To our knowledge, no previous work has targeted DAM genes in perennial fruit trees to alter dormancy. However, there is a known, natural DAM evergrowing (evg) peach mutant which is the result of a genomic deletion of four DAM genes and which does not enter dormancy when exposed to low temperatures or shortening days [231]. More research is needed to determine the effects of targeted DAM mutagenesis on dormancy in pome fruits and its applicability in commercial fruit production. Also, alternative targets need to be identified to more precisely fine tune chilling requirements.
It is predicted that winter warming and resulting disturbed dormancy will be an issue for temperate fruit production in increasingly larger areas of the world [232]. For example, due to rising winter temperatures in Japan, flowering disorders occur more frequently in P. pyrifolia “Hosui” trees which as a result show erratic flowering, asynchronous bud-break and bud loss due to inadequate chilling during the dormancy phase [223]. However, increased temperatures are not always negative for pome fruit production. For example, European pear (P. communis) production of the cultivar “Conference” in Belgium is predicted to be at a lower risk of frost damage-related production losses because of the decreased occurrence of frost days during the flowering period [233]. These two examples show that the consequences of climate change on pome fruit phenology trees are complex and region- and cultivar-specific. In given examples, a decreased chilling requirement may improve uniform bud break in “Hosui” pears grown in Japan, but could expose “Conference” pears grown in Belgium to frost damage due to early flowering. So, when adapting pome fruit tree phenology by breeding or gene editing, it is recommended to obtain an accurate view of the specific challenges present in the crop and region of interest.
3.5.3 Biennial Bearing
Biennial bearing (BB) occurs when a fruit tree has an alternating pattern of low and high fruit production over consecutive years. This is caused by the inhibition of flower induction (FI) in the meristems by growing fruits, since FI occurs simultaneously with fruit development during 4–8 weeks after full bloom [234]. In pome fruit trees, each individual spur is biannual and can only fruit every other year, commonly named ‘ON’ and ‘OFF’ years. During ‘ON’ years there is abundant flowering and potentially a heavy fruit set, followed by an ‘OFF’-year with mostly vegetative growth and thus less potential for fruit set. Ideally, a fruit tree will have a balanced proportion of ‘OFF’ spurs and ‘ON’ spurs each year, resulting in a predictable and constant fruit production. If a certain event triggers low fruit set or causes early loss of fruitlets, all spurs on a tree will be synchronized to an ‘OFF’ status and, consequently, an ‘ON’ status next year. This starts a cycle of BB which causes variability in yield and fruit size over the years and is therefore undesired by the grower. A frequently applied measure to avoid the continuation of the BB cycle is early crop thinning in ‘ON’ years by removing flowers or young fruitlets. This can be done chemically by using compounds that damage flower organs and so inhibit fruit set or by mechanically removing the young fruitlets [235, 236]. During ‘OFF’ years gibberellic acids such as GA7 can be applied to the trees after bloom to repress excess FI [237]. In addition to crop thinning, optimal pruning of the fruit trees will maintain young spurs which are less susceptible to BB than older spurs [235]. But despite these precautions and good management, there are still differences in susceptibility of different pome fruit cultivars to BB, especially in apple. This implies that there is a genetic basis determining susceptibility or resistance to BB in certain cultivars [238].
BB was long thought to be the result of hormonal signaling from the developing fruits to nearby developing buds. In 1998, diffusible auxins were shown to be present in the seeds of developing fruits during the period of FI, and the levels of these auxins in fruits increased with an increased number of seeds [239]. A recent study compared the apple cultivars “Gala” (a regular bearer) and “Fuji” (a biennial bearer) and found that “Fuji” had a higher average seed number per fruit compared to “Gala” [240]. The “Fuji” fruits did not only have a higher number of seeds per fruit, each seed also had higher levels of cytokinins and auxins than seeds from “Gala” fruits. On the contrary, “Gala” seeds had increased levels of GA3 and GA19 compared to “Fuji”. These plant hormones were also found to be exported by diffusion through the stem of the apple fruit in both cultivars. These differences in hormone production in seeds could explain the susceptibility of “Fuji” to BB, since some GAs repress FI in apple trees trough currently unknown mechanisms [241]. Several QTL’s were identified for biennial bearing in apple, some included flowering genes like BFTa, SOC1-like and COL1, others included hormonal factors such as genes involved in GA biosynthesis GIBBERELLIN 2-OXIDASE (GA2ox), GIBBERELLIN 20-OXIDASE (GA20ox) and GA3ox-like-b and auxin related genes such as AFB6 [242].
A study in “Gala” apple trees identified many differentially expressed genes (DEGs) between ‘ON’ and ‘OFF’ buds in trees by artificially inducing biennial bearing through manual flower removal during bloom. The authors found many DEGs among the flowering genes, including flowering repressor genes such as TEMPRANILLO1 (TEM1) and MAF2 (MADS AFFECTING FLOWERING 2). Also flowering promoting transcription factors, SQUAMOSA PROMOTER BINDING-LIKE 5 (SPL5) and SPL9 and FLORAL TRANSITION AT MERISTEM (FTM1) were upregulated in ‘OFF’ trees, simultaneously with APELLATA1 (AP1) genes. Additionally, secondary metabolism genes were downregulated in the shoot apical meristem (SAM) of ‘ON’ trees, possibly due to the developing fruits causing carbohydrate depletion in the SAM. This downregulation of secondary metabolism genes is an indication of reduced cell division in the apical tissues and is supported by the observed up-regulation of KNOTTED-LIKE FROM ARABIDOPSIS THALIANA (KNAT1) and NO APICAL MERISTEM (NAM) genes during ‘ON’ years. KNAT1 and NAM are both genes which prevent meristematic tissue differentiation and thus maintain the SAM, preventing FI and differentiation of floral tissues under low-carbon conditions. Also axillary meristem (AM) regulating genes were upregulated in ‘ON’ buds, such as MORE AXILLARY BRANCHES 1 (MAX1) and BRANCHED1 (BRC1), inhibiting AM formation and axillary bud outgrowth, respectively [243, 244]. This reduction in AM formation and axillary bud outgrowth results in reduced vegetative growth during ‘ON’ years with heavy fruit bearing. The study also found DEGs related to auxin, abscisic acid, brassinosteroid and ethylene. The Gibberellic Acid (GA) biosynthesis genes GA2ox and GA20ox were shown to be upregulated in ‘ON’ years, confirming the previously mentioned studies that showed these genes as QTLs for BB [242, 244]. This is not surprising, since GA treatments during ‘OFF’ years are used in horticulture to inhibit FI [245].
Also epigenetic modifications are involved in regulation of BB, as it was shown that differentially methylated regions (DMRs) are present between ‘ON’ and ‘OFF’ trees in buds of the apple cultivar “Fuji” [246]. Many flowering genes showed to be differentially methylated such as MADS-box, COL, B-box, NFY and SPL. Also genes involved in hormonal signaling such as gibberellin, auxin and jasmonic acid showed to be DMRs, again highlighting the importance of hormonal regulation of FI in BB in apple.
Differences in gene expression in developing buds during ‘ON’ and ‘OFF’ years are very valuable in understanding FI and the genes involved, but it does not provide evidence to identify the genetic factors responsible for susceptibility of a cultivar to BB. Since the exact molecular mechanisms of BB are not yet well characterized, it is difficult to put forward a good candidate gene for CRISPR gene editing. Since all genes that were identified so far in BB are involved in crucial plant functions as flowering and hormonal signaling, the generation of knock-out (KO) mutants may not be a suitable approach due to the possible occurrence of undesirable side effects in tree phenology and/or flower morphology [247]. A better understanding of the genetic control of BB is needed to apply innovative gene editing techniques such as CRISPR base editing. Since there are observations that GA and auxin production in the seeds play a role in BB, CRISPR transcriptional activation could be used to mimic expression patterns of hormone biosynthesis genes of resistant cultivars in susceptible cultivars.
4 Concluding Remarks
Apple and pear are among the most cultivated temperate fruit crops, but the threats to their production remain significant. Many of the currently grown cultivars still suffer from high susceptibility to pests, diseases and abiotic stress, low fruit quality, biennial bearing, suboptimal yields and other problems. Effective breeding strategies are necessary to tackle these problems. However, current breeding approaches are too time-consuming and expensive to profitably develop improved cultivars and to meet the rapidly evolving breeding requirements, especially in light of impending climate change. In addition, apple and pear production are traditionally dominated by just a handful of cultivars with a high market recognition, such as “Gala”, “Golden Delicious” and “Fuji” for apple, and “Conference”, “Bartlett” and “Doyenné du Comice” for pear. It has proven difficult to introduce new cultivars in the market, therefore, enhancing important traits in established cultivars is a very effective strategy, albeit extremely difficult using conventional breeding techniques. There is therefore a great need for faster and more accurate breeding approaches.
The recent advancement of gene-editing techniques such as CRISPR/Cas related technologies can help address these issues. Although successful gene-editing applications in pome fruit are still limited compared to many model crops, several proof-of-concept studies in apple, pear and some other perennial tree species have shown that gene-editing is feasible in pome fruits and that there are numerous potential breeding applications. This chapter discussed the current state-of-the-art, challenges and opportunities regarding gene-editing in pome fruit, as well as potential gene-targets for new applications in light of important breeding objectives. Several technical challenges remain to be solved in order to successfully apply gene-editing in pome fruit breeding including the high recalcitrant nature of pome fruit plantlets to in vitro propagation and regeneration, the low transformation efficiencies, and the complex removal of transgenes by crossing due to the long life cycle and high heterozygosity of pome fruit tree species. Improvements to the original gene-editing approaches that are tailored to the specific problems in pome fruit and other perennial tree species will help improve the application potential in pome fruit breeding.
References
FAO: World Food and Agriculture – Statistical Yearbook 2021. ROME (2021)
FAOSTAT, FAO: Crops and livestock products. License:CC BY-NC-SA 3.0 IGO (2023)
Penna, S., Jain, S.M.: Fruit crop improvement with genome editing, in vitro and transgenic approaches. Horticulturae. 9(58), 1–20 (2023)
Kumar, S., Kirk, C., Deng, C., Wiedow, C., Knaebel, M., Brewer, L.: Genotyping-by-sequencing of pear (Pyrus spp.) accessions unravels novel patterns of genetic diversity and selection footprints. Hortic. Res. 4, 17015 (2017)
Saito, T.: Advances in Japanese pear breeding in Japan. Breed. Sci. 66(1), 46–59 (2016)
Li, J., et al.: Pear genetics: recent advances, new prospects, and a roadmap for the future. Hortic. Res. 9, uhab040 (2022)
Kumar, S., Hilario, E., Deng, C.H., Molloy, C.: Turbocharging introgression breeding of perennial fruit crops: a case study on apple. Hortic. Res. 7, 47 (2020a)
Chagné, D., et al.: The draft genome sequence of European pear (Pyrus communis L. ‘Bartlett’). PLoS One. 9(4), e92644 (2014)
Velasco, R., et al.: The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 42(10), 833–839 (2010)
Montanari, S., et al.: Development of a highly efficient Axiom™ 70 K SNP array for Pyrus and evaluation for high-density mapping and germplasm characterization. BMC Genomics. 20(1), 1–18 (2019)
Chagné, D., et al.: Validation of SNP markers for fruit quality and disease resistance loci in apple (Malus × domestica Borkh.) using the OpenArray® platform. Hortic. Res. 6, 30 (2019)
Alvarez, D., et al.: Fruit crops in the era of genome editing: closing the regulatory gap. Plant Cell Rep. 40(6), 915–930 (2021)
Peer, R., et al.: Targeted mutagenesis using zinc-finger nucleases in perennial fruit trees. Planta. 241(4), 941–951 (2015)
Wyman, C., Kanaar, R.: DNA double-strand break repair: All’s well that ends well. Annu. Rev. Genet. 40, 363–383 (2006)
Kim, S., Kim, J.S.: Targeted genome engineering via zinc finger nucleases. Plant Biotechnol. Rep. 5(1), 9–17 (2011)
Kim, Y.G., Cha, J., Chandrasegaran, S.: Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U. S. A. 93(3), 1156–1160 (1996)
Ramirez-Torres, F., et al.: Genome editing in fruit, ornamental, and industrial crops. Transgenic Res. 30(4), 499–528 (2021)
Bhoi, A., Yadu, B., Chandra, J., Keshavkant, S.: Mutagenesis: A coherent technique to develop biotic stress resistant plants. Plant Stress. 3, 100053 (2022)
Osakabe, K., Osakabe, Y., Toki, S.: Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc. Natl. Acad. Sci. U. S. A. 107(26), 12034–12039 (2010)
Lloyd, A., Plaisier, C., Carroll, D., Drews, G.: Targeted mutagenesis in Arabidopsis using zinc-finger nucleases. Proc. Natl. Acad. Sci. 102(6), 2232–2237 (2004)
Curtin, S.J., et al.: Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol. 156(2), 466–473 (2011)
Jung, Y.J., Nogoy, F.M., Lee, S.K., Cho, Y.G., Kang, K.K.: Application of ZFN for site directed mutagenesis of Rice SSIVa gene. Biotechnol. Bioprocess Eng. 23(1), 108–115 (2018)
Lu, H., Klocko, A.L., Dow, M., Ma, C., Amarasinghe, V., Strauss, S.H.: Low frequency of zinc-finger nuclease-induced mutagenesis in Populus. Mol. Breed. 36(9), 1–13 (2016)
Zhang, Y., Massel, K., Godwin, I.D., Gao, C.: Applications and potential of genome editing in crop improvement. Genome Biol. 19(1), 1–11 (2018)
Christian, M., et al.: Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 186(2), 756–761 (2010)
Son, S., Park, S.R.: Challenges facing CRISPR/Cas9-based genome editing in plants. Front. Plant Sci. 13(902413), 1–20 (2022)
Bortesi, L., Fischer, R.: The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33(1), 41–52 (2015)
Li, T., Liu, B., Spalding, M.H., Weeks, D.P., Yang, B.: High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30(5), 390–392 (2012a)
Shan, Q., Zhang, Y., Chen, K., Zhang, K., Gao, C.: Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol. J. 13(6), 791–800 (2015)
Wang, Y., et al.: Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32(9), 947–951 (2014)
Char, S.N., et al.: Heritable site-specific mutagenesis using TALENs in maize. Plant Biotechnol. J. 13(7), 1002–1010 (2015)
Kannan, B., Jung, J.H., Moxley, G.W., Lee, S.M., Altpeter, F.: TALEN-mediated targeted mutagenesis of more than 100 COMT copies/alleles in highly polyploid sugarcane improves saccharification efficiency without compromising biomass yield. Plant Biotechnol. J. 16(4), 856–866 (2018)
Jung, J.H., Altpeter, F.: TALEN mediated targeted mutagenesis of the caffeic acid O-methyltransferase in highly polyploid sugarcane improves cell wall composition for production of bioethanol. Plant Mol. Biol. 92(1–2), 131–142 (2016)
Nishitani, C., et al.: Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 6, 31481 (2016)
Jiang, F., Doudna, J.A.: CRISPR – Cas9 structures and mechanisms. Annu. Rev. Biophys. 46, 505–529 (2017)
Qin, G., et al.: Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 17(5), 471–482 (2007)
Pompili, V., Dalla Costa, L., Piazza, S., Pindo, M., Malnoy, M.: Reduced fire blight susceptibility in apple cultivars using a high-efficiency CRISPR/Cas9-FLP/FRT-based gene editing system. Plant Biotechnol. J. 18(3), 845–858 (2020)
Charrier, A., Vergne, E., Dousset, N., Richer, A., Petiteau, A., Chevreau, E.: Efficient targeted mutagenesis in apple and first time edition of pear using the CRISPR-Cas9 system. Front. Plant Sci. 10(40), 1–12 (2019)
Zhang, Y., Zhou, P., Bozorov, T.A., Zhang, D.: Application of CRISPR/Cas9 technology in wild apple (Malus sieverii) for paired sites gene editing. Plant Methods. 17(1), 1–9 (2021a)
Schröpfer, S., Flachowsky, H.: Tracing CRISPR/Cas12a mediated genome editing events in apple using high-throughput genotyping by PCR capillary gel electrophoresis. Int. J. Mol. Sci. 22, 12611 (2021)
Zetsche, B., Gootenberg, J.S., Abudayyeh, O.O., Slaymaker, I.M., Makarova, K.S., Essletzbichler, P., Volz, S.E., et al.: Cpf1 Is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 163, 759–771 (2015)
An, Y., et al.: Efficient genome editing in populus using CRISPR/Cas12a. Front. Plant Sci. 11, 1–9 (2020)
Jia, H., Orbović, V., Wang, N.: CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnol. J. 17(10), 1928–1937 (2019)
Bandyopadhyay, A., Kancharla, N., Javalkote, V.S., Dasgupta, S., Brutnell, T.P.: CRISPR-Cas12a (Cpf1): a versatile tool in the plant genome editing tool box for agricultural advancement. Front. Plant Sci. 11, 584151 (2020)
Malabarba, J., Chevreau, E., Dousset, N., Veillet, F., Moizan, J., Vergne, E.: New strategies to overcome present CRISPR/Cas9 limitations in apple and pear: efficient Dechimerization and base editing. Int. J. Mol. Sci. 22, 319 (2021)
Komor, A.C., Kim, Y.B., Packer, M.S., Zuris, J.A., Liu, D.R.: Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 533(7603), 420–424 (2016)
Reed, K.M., Bargmann, B.O.R.: Protoplast regeneration and its use in new plant breeding technologies. Front. Genome Ed. 3, 734951 (2021)
Osakabe, Y., et al.: CRISPR–Cas9-mediated genome editing in apple and grapevine. Nat. Protoc. 13(12), 2844–2863 (2018)
Malnoy, M., et al.: DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7, 1904 (2016)
Chen, J., et al.: Significant improvement of apple (Malus domestica Bork.) transgenic plant production by pre-transformation with a Baby boom transcription factor. Hortic. Res. 9, uhab014 (2022)
Lobato-Gómez, M., Hewitt, S., Capell, T., Christou, P., Dhingra, A., Girón-Calva, P.S.: Transgenic and genome-edited fruits: background, constraints, benefits, and commercial opportunities. Hortic. Res. 8, 166 (2021)
Voss-Fels, K.P., Stahl, A., Hickey, L.T.: Q&A: modern crop breeding for future food security. BMC Biol. 17(1), 18 (2019)
Lakso, A., Robinson, T.: Sunlight, yield, and productivity of apples. Tree Physiol. 117(4), 551–557 (2013)
Anthony, B., Serra, S., Musacchi, S.: Optimizing crop load for new apple cultivar: “WA38”. Agronomy. 9, 107 (2019)
Devoghalaere, F., et al.: A genomics approach to understanding the role of auxin in apple (Malus x domestica) fruit size control. BMC Plant Biol. 12, 7 (2012)
Liu, W., et al.: Research progress on genetic basis of fruit quality traits in apple (Malus × domestica). Front. Plant Sci. 13, 918202 (2022)
Theron, K.I.: Size matters: Factors influencing fruit size in pear. Acta Hortic. 909, 545–556 (2011)
Subbaraya, U., Rajendran, S., Simeon, S., Suthanthiram, B., Marimuthu Somasundram, S.: Unravelling the regulatory network of transcription factors in parthenocarpy. Sci. Hortic. (Amsterdam). 261, 108920 (2020)
Dang, Q., Sha, H., Nie, J., Wang, Y., Yuan, Y., Jia, D.: An apple (Malus domestica) AP2/ERF transcription factor modulates carotenoid accumulation. Hortic. Res. 8(223), 1–12 (2021)
Mignard, P., Beguería, S., Giménez, R., Font I Forcada, C., Reig, G., Moreno, M.Á.: Effect of genetics and climate on apple sugars and organic acids profiles. Agronomy. 12(4), 827 (2022)
Amyotte, B., Bowen, A.J., Banks, T., Rajcan, I., Somers, D.J.: Mapping the sensory perception of apple using descriptive sensory evaluation in a genome wide association study. PLoS One. 12(2), e0171710 (2017)
Jung, M., et al.: Genetic architecture and genomic predictive ability of apple quantitative traits across environments. Hortic. Res. 9, uhac028 (2022)
Kumar, S., et al.: GWAS provides new insights into the genetic mechanisms of phytochemicals production and red skin colour in apple. Hortic. Res. 9, uhac218 (2022)
Mcclure, K.A., et al.: A genome-wide association study of apple quality and scab resistance. Plant Genome. 11(1), 170075 (2018)
Ikegaya, A., Toyoizumi, T., Ohba, S., Nakajima, T., Kawata, T., Ito, S., Arai, E.: Effects of distribution of sugars and organic acids on the taste of strawberries. Food Sci. Nutr. 7, 2419–2426 (2019)
Aprea, E., et al.: Sweet taste in apple: the role of sorbitol, individual sugars, organic acids and volatile compounds. Nat. Publ. Gr. 7, 44950 (2017a)
Larsen, B., et al.: Genome-wide association studies in apple reveal loci for aroma volatiles, sugar composition, and harvest date. Plant Genome. 12(2), 180104 (2019)
Nishio, S., et al.: Genome-wide association study of individual sugar content in fruit of Japanese pear (Pyrus spp.). BMC Plant Biol. 21(1), 378 (2021)
Guan, Y., et al.: QTLs detected for individual sugars and soluble solids content in apple. Mol. Breed. 35(135), 1–13 (2015)
Miller, B.A., Kostick, S.A., Luby, J.J.: Large-effect QTLs for titratable acidity and soluble solids content validated in ‘honeycrisp’-derived apple germplasm. Agronomy. 12(7), 1703 (2022)
Kyung Hyun, T., Hee Eom, S., Kim, J.-S.: Genomic analysis and gene structure of the two invertase families in the domesticated apple (Malus × domestica Borkh.). Plant Omi. J. 4(7), 391–399 (2011)
Li, M., Feng, F., Cheng, L.: Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development. PLoS One. 7(3), 33055 (2012b)
Jiang, S., Li, S., Luo, J., Wang, X., Shi, C.: QTL mapping and transcriptome analysis of sugar content during fruit ripening of Pyrus pyrifolia. Front. Plant Sci. 14, 13 (2023)
Zhen, Q., et al.: Developing gene-tagged molecular markers for evaluation of genetic association of apple SWEET genes with fruit sugar accumulation. Hortic. Res. 5, 14 (2018)
Nie, P., Xu, G., Yu, B., Lyu, D., Xue, X., Qin, S.: Genome-wide identification and expression profiling reveal the potential functions of the SWEET gene family during the sink organ development period in apple (Malus × domestica Borkh.). Agronomy. 12(8), 1747 (2022)
Ma, B., et al.: Determination of predominant organic acid components in Malus species: correlation with apple domestication. Meta. 8(74), 1–20 (2018)
Zhang, Y., Li, P., Cheng, L.: Developmental changes of carbohydrates, organic acids, amino acids, and phenolic compounds in “Honeycrisp” apple flesh. Food Chem. 123(4), 1013–1018 (2010)
Wu, J., et al.: Chemical compositional characterization of some apple cultivars. Food Chem. 103(1), 88–93 (2007)
Aprea, E., et al.: Sweet taste in apple: the role of sorbitol, individual sugars, organic acids and volatile compounds. Nat. Publ. Gr. 7(44950), 1–11 (2017b)
Xu, K., Wang, A., Brown, S.: Genetic characterization of the Ma locus with pH and titratable acidity in apple. Mol. Breed. 30, 899–912 (2012)
Kenis, K., Keulemans, J., Davey, M.W.: Identification and stability of QTLs for fruit quality traits in apple. Tree Genet. Genomes. 4(4), 647–661 (2008)
Ma, B., et al.: Genes encoding aluminum-activated malate transporter II and their association with fruit acidity in apple. Plant Genome. 8(3), 1–20 (2015)
Ban, S., Xu, K.: Identification of two QTLs associated with high fruit acidity in apple using pooled genome sequencing analysis. Hortic. Res. 7, 171 (2020)
Visser, T., Verhaegh, J.J.: Inheritance and selection of some fruit characters of apple. I. Inheritance of low and high acidity. Euphytica. 27(3), 753–760 (1978)
Liebhard, R., Koller, B., Gianfranceschi, L., Gessler, C.: Creating a saturated reference map for the apple (Malus x domestica Borkh.) genome. Theor. Appl. Genet. 106(8), 1497–1508 (2003)
Verma, S., et al.: Two large-effect QTLs, Ma and Ma3, determine genetic potential for acidity in apple fruit: breeding insights from a multi-family study. Tree Genet. Genomes. 15(18), 1–20 (2019)
Maliepaard, C., et al.: Aligning male and female linkage maps of apple (Malus pumila Mill.) using multi-allelic markers. Theor. Appl. Genet. 97, 60–73 (1998)
Kumar, S., Chagné, D., Bink, M., Volz, R.K., Whitworth, C., Carlisle, C.: Genomic selection for fruit quality traits in apple (Malus×domestica Borkh.). PLoS One. 7(5), e36674 (2012)
Bai, Y., Dougherty, L., Li, M., Fazio, G., Cheng, L., Xu, K.: A natural mutation-led truncation in one of the two aluminum-activated malate transporter-like genes at the Ma locus is associated with low fruit acidity in apple. Mol. Gen. Genomics. 287(8), 663–678 (2012)
Li, C., et al.: Apple ALMT9 requires a conserved C-terminal domain for malate transport underlying fruit acidity. Plant Physiol. 182, 992–1006 (2020)
Hu, D.G., Sun, C.-H., Ma, Q.-J., You, C.-X., Cheng, L., Hao, Y.-J.: MdMYB1 regulates anthocyanin and malate accumulation by directly facilitating their transport into vacuoles in apples. Plant Physiol. 170, 1315–1330 (2016)
Hu, D.G., et al.: The R2R3-MYB transcription factor MdMYB73 is involved in malate accumulation and vacuolar acidification in apple. Plant J. 91(3), 443–454 (2017)
Zhang, Q.-Y., et al.: BTB-BACK-TAZ domain protein MdBT2-mediated MdMYB73 ubiquitination negatively regulates malate accumulation and vacuolar acidification in apple. Hortic. Res. 7(151), 1–11 (2020)
Jia, D., et al.: Genetic variation in the promoter of an R2R3−MYB transcription factor determines fruit malate content in apple (Malus domestica Borkh.). Plant Physiol. 186(1), 549 (2021)
Jia, D., et al.: Apple fruit acidity is genetically diversified by natural variations in three hierarchical epistatic genes: Mdsaur37, mdpp2ch and mdalmtii. Plant J. 95(3), 427–443 (2018a)
Farneti, B., et al.: Is there room for improving the nutraceutical composition of apple? J. Agric. Food Chem. 63, 2750–2759 (2015)
Davies, T., Watts, S., McClure, K., Migicovsky, Z., Myles, S.: Phenotypic divergence between the cultivated apple (Malus domestica) and its primary wild progenitor (Malus sieversii). PLoS One. 17(3), e0250751 (2022)
McClure, K.A., et al.: Genome-wide association studies in apple reveal loci of large effect controlling apple polyphenols. Hortic. Res. 6(1), 107 (2019)
Bilbrey, E.A., Williamson, K., Hatzakis, E., Miller, D.D., Fresnedo-Ramírez, J., Cooperstone, J.L.: Integrating genomics and multiplatform metabolomics enables metabolite quantitative trait loci detection in breeding-relevant apple germplasm. New Phytol. 232(5), 1944–1958 (2021)
Khan, S.A., et al.: Genetic analysis of metabolites in apple fruits indicates an mQTL hotspot for phenolic compounds on linkage group 16. J. Exp. Bot. 63(8), 2895–2908 (2012)
Gosch, C., et al.: Substrate specificity and contribution of the glycosyltransferase UGT71A15 to phloridzin biosynthesis. Trees Struct. Funct. 26(1), 259–271 (2012)
Miranda, S., Piazza, S., Nuzzo, F., Li, M., Lagrèze, J., Mithöfer, A., Cestaro, A., et al.: CRISPR/Cas9 genome-editing applied to MdPGT1 in apple results in reduced foliar phloridzin without impacting plant growth. Plant J. 113, 92–105 (2023)
Espley, R.V., Hellens, R.P., Putterill, J., Stevenson, D.E., Kutty-Amma, S., Allan, A.C.: Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427 (2007)
Xu, H., et al.: MdMYB6 regulates anthocyanin formation in apple both through direct inhibition of the biosynthesis pathway and through substrate removal. Hortic. Res. 7, 72 (2020)
Wang, S., et al.: Apple MdMYB306-like inhibits anthocyanin synthesis by directly interacting with MdMYB17 and MdbHLH33. Plant J. 110(4), 1021–1034 (2022)
Colanero, S., Perata, P., Gonzali, S.: The atroviolacea gene encodes an R3-MYB protein repressing anthocyanin synthesis in tomato plants. Front. Plant Sci. 9, 830 (2018)
Davuluri, G.R., et al.: Manipulation of DET1 expression in tomato results in photomorphogenic phenotypes caused by post-transcriptional gene silencing. Plant J. 40(3), 1–15 (2004)
Valpuesta, V., Botella, M.A.: Biosynthesis of L-ascorbic acid in plants: new pathways for an old antioxidant. Trends Plant Sci. 9(12), 573–577 (2004)
Zheng, X., et al.: Metabolism and regulation of ascorbic acid in fruits. Plants. 11(12), 1602 (2022)
Mellidou, I., Chagné, D., Laing, W.A., Keulemans, J., Davey, M.W.: Allelic variation in paralogs of GDP-l-galactose phosphorylase is a major determinant of vitamin C concentrations in apple fruit. Plant Physiol. 160(3), 1613 (2012)
Khan, A., Korban, S.S.: Breeding and genetics of disease resistance in temperate fruit trees: challenges and new opportunities. Theor. Appl. Genet. 135(11), 3961–3985 (2022)
Kellerhals, M., et al.: European pome fruit genetic resources evaluated for disease resistance. Trees Struct. Funct. 26(1), 179–189 (2012)
Zhao, Y.Q., et al.: Fire blight disease, a fast-approaching threat to apple and pear production in China. J. Integr. Agric. 18(4), 815–820 (2019)
Höfer, M., Flachowsky, H., Schröpfer, S., Peil, A.: Evaluation of scab and mildew resistance in the gene bank collection of apples in Dresden-pillnitz. Plan. Theory. 10(6), 1–16 (2021)
Papp, D., Singh, J., Gadoury, D., Khan, A.: New North American isolates of Venturia inaequalis can overcome apple scab resistance of Malus floribunda 821. Plant Dis. 104(3), 649–655 (2020)
Fahrentrapp, J., et al.: A candidate gene for fire blight resistance in Malus × robusta 5 is coding for a CC–NBS–LRR. Tree Genet. Genomes. 9, 237–251 (2013)
Emeriewen, F., Richter, K., Flachowksy, H., Malnoy, M., Peil, A.: Genetic analysis and fine mapping of the fire blight resistance locus of Malus × arnoldiana on linkage group 12 reveal first candidate genes. Front. Plant Sci. 12, 667133 (2021)
Durel, C.E., Denancé, C., Brisset, M.N.: Two distinct major QTL for resistance to fire blight co-localize on linkage group 12 in apple genotypes “Evereste” and Malus floribunda clone. Genome. 52, 139–147 (2009)
Emeriewen, O.F., et al.: Towards map-based cloning of FB_Mfu10: identification of a receptor-like kinase candidate gene underlying the Malus fusca fire blight resistance locus on linkage group 10. Mol. Breed. 38, 106 (2018)
Patocchi, A., et al.: Ten years of VINQUEST: first insight for breeding new apple cultivars with durable apple scab resistance. Plant Dis. 104(8), 2074–2081 (2020)
Shoda, S.T.M., Gonai, Y.A.T., Sawamura, M.K.Y., Kotobuki, H.I.K., Gessler, A.P.C., Yamamoto, T.H.T.: Genetic mapping of the pear scab resistance gene Vnk of Japanese pear cultivar Kinchaku. Theor. Appl. Genet. 113, 743–752 (2006)
Cho, K.H., Shin, I.S., Kim, K.T., Suh, E.J., Hong, S.S., Lee, H.J.: Development of AFLP and CAPS markers linked to the scab resistance gene, Rvn2, in an inter-specific hybrid pear (Pyrus spp.). J. Hortic. Sci. Biotechnol. 84(6), 619–624 (2015)
Oh, S., Han, H., Kim, D.: A novel pear scab (Venturia nashicola) resistance gene, Rvn3, from interspecific hybrid pear (Pyrus pyrifolia × P. communis). Plants. 10, 2632 (2021)
Bouvier, L., et al.: A new pear scab resistance gene Rvp1 from the European pear cultivar “Navara” maps in a genomic region syntenic to an apple scab resistance gene cluster on linkage group 2. Tree Genet. Genomes. 8, 53–60 (2012)
Caffier, V., Laurens, F.: Breakdown of Pl2, a major gene of resistance to apple powdery mildew, in a French experimental orchard. Plant Pathol. 54, 116–124 (2005)
Kost, T.D., Gessler, C., Jänsch, M., Flachowsky, H., Patocchi, A., Broggini, G.A.L.: Development of the first cisgenic apple with increased resistance to fire blight. PLoS One. 10(12), e0143980 (2015)
Broggini, G., et al.: Engineering fire blight resistance into the apple cultivar “Gala” using the FB_MR5 CC-NBS-LRR resistance gene of. Plant Biotechnol. 12, 728–733 (2014)
Perchepied, L., et al.: Successful intergeneric transfer of a major apple scab resistance gene (Rvi6) from apple to pear and precise comparison of the downstream molecular mechanisms of this resistance in both species. BMC Genomics. 22, 843 (2021)
Campa, M., et al.: HIPM is a susceptibility gene of Malus spp.: reduced expression reduces susceptibility to Erwinia amylovora. Mol. Plant-Microbe Interact. 32(2), 167–175 (2019)
Malnoy, M., Meng, X., Bonasera, J.M., Nissinen, R.M.: Silencing of apple proteins that interact with DspE, a pathogenicity effector from Erwinia amylovora, as a strategy to increase resistance to fire blight. Acta Hortic. 663, 469–474 (2004)
Pessina, S., et al.: The knock-down of the expression of MdMLO19 reduces susceptibility to powdery mildew (Podosphaera leucotricha) in apple (Malus domestica). Plant Biotechnol. J. 14, 2033–2044 (2016)
Kusch, S., Panstruga, R.: mlo-based resistance: an apparently universal “weapon” to defeat powdery mildew disease. Mol. Plant-Microbe Interact. 30(3), 179–189 (2017)
Åhman, I., Kim, S., Zhu, L.: Plant genes benefitting aphids — potential for exploitation in resistance breeding. Front. Plant Sci. 10, 1452 (2019)
Kloth, K.J., et al.: AtWRKY22 promotes susceptibility to aphids and modulates salicylic acid and jasmonic acid signalling. J. Exp. Bot. 67(11), 3383–3396 (2016)
Kyrou, K., et al.: Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36(11), 1062–1066 (2018)
Paul, N.C., et al.: Plant and fungal genome editing to enhance plant disease resistance using the CRISPR / Cas9 system. Front. Plant Sci. 12, 700925 (2021)
Burt, A.: Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. R. Soc. B Biol. Sci. 270(1518), 921–928 (2003)
Bier, E.: Gene drives gaining speed. Nat. Rev. Genet. 23(1), 5–22 (2022)
Tek, M.I., Budak, K.: A new approach to develop resistant cultivars against the plant pathogens: CRISPR drives. Front. Plant Sci. 13, 889497 (2022)
Gardiner, D.M., Rusu, A., Barrett, L., Hunter, G.C., Kazan, K.: Can natural gene drives be part of future fungal pathogen control strategies in plants? New Phytol. 228(4), 1431–1439 (2020)
Graeff, N., DeJongsma, K.R., Bredenoord, A.L.: Experts’ moral views on gene drive technologies: a qualitative interview study. BMC Med. Ethics. 22(25), 1–15 (2021)
Jiang, L., et al.: MdGH3.6 is targeted by MdMYB94 and plays a negative role in apple water-deficit stress tolerance. Plant J. 109, 1271–1289 (2022a)
Jiang, L., et al.: Engineering drought-tolerant apple by knocking down six GH3 genes and potential application of transgenic apple as a rootstock. Hortic. Res. 9, uhac122 (2022b)
Clemens, M., et al.: VvEPFL9-1 Knock-out via CRISPR / Cas9 reduces stomatal density in grapevine. Front. Plant Sci. 13, 878001 (2022)
Kumar, R., Suhas, J., Bharat, S., Mishra, R.: Engineering drought tolerance in plants through CRISPR / Cas genome editing. Biotech. 10, 400 (2020b)
Osakabe, Y., Watanabe, T., Sugano, S.S., Ueta, R., Ishihara, R.: Optimization of CRISPR / Cas9 genome editing to modify abiotic stress responses in plants. Nat. Publ. Gr. 6, 26685 (2016)
Salazar-gutiérrez, M.R., Chaves, B., Hoogenboom, G.: Scientia Horticulturae Freezing tolerance of apple flower buds. Sci. Hortic. (Amsterdam). 198, 344–351 (2016)
Shen, X., et al.: The RNA-binding protein MdHYL1 modulates cold tolerance and disease resistance in apple. Plant Physiol. 17, 1–18 (2023)
Liang, Y., et al.: Transcriptional regulation of bark freezing tolerance in apple (Malus domestica Borkh.). Hortic. Res. 7, 205 (2020)
Locatelli, F., Baldoni, Æ.E., Mattana, Æ.M.: Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Rep. 27, 1677–1686 (2008)
Bai, T., Zheng, X.: Genome-wide characterization of the HSP20 gene family identifies potential members involved in temperature stress response in apple. Front. Genet. 11, 609184 (2020)
Torres, C.A., Sepúlveda, G., Kahlaoui, B.: Phytohormone interaction modulating fruit responses to photooxidative and heat stress on apple (Malus domestica Borkh.). Front. Plant Sci. 8, 2129 (2017)
Zhang, X., et al.: Genome-wide identification of PRP genes in apple genome and the role of MdPRP6 in response to heat stress. Int. J. Mol. Sci. 22, 5942 (2021b)
Dong, Q., et al.: MdVQ37 overexpression reduces basal thermotolerance in transgenic apple by affecting transcription factor activity and salicylic acid homeostasis. Hortic. Res. 8, 220 (2021)
Zhu, J.: Abiotic stress signaling and responses in plants. Cell. 167, 313–324 (2016)
Hu, L., et al.: MdINT1 enhances apple salinity tolerance by regulating the antioxidant system, homeostasis of ions, and osmosis. Plant Physiol. Biochem. 154, 689–698 (2020)
Chen, S., et al.: Knockout of the entire family of AITR genes in Arabidopsis leads to enhanced drought and salinity tolerance without fitness costs. BMC Plant Biol. 21, 137 (2021)
Zhu, Y., et al.: An Arabidopsis Nucleoporin NUP85 modulates plant responses to ABA and salt stress. Plos Genet. 13(12), e1007124 (2017)
Zhang, A., Liu, Y., Wang, F., Li, T., Chen, Z., Kong, D.: Enhanced rice salinity tolerance via CRISPR / Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 39, 47 (2019)
Bouzroud, S., et al.: Down regulation and loss of Auxin Response Factor 4 function using CRISPR/Cas9 alters plant growth, stomatal function and improves tomato tolerance to salinity and osmotic stress. Genes (Basel). 11, 272 (2020)
Okada, K., Honda, C.: Molecular mechanisms regulating the columnar tree architecture in apple. Forests. 13, 1084 (2022)
Hill, J.L., Hollender, C.A.: Branching out: new insights into the genetic regulation of shoot architecture in trees. Curr. Opin. Plant Biol. 47, 73–80 (2019)
Bryant, D.W., et al.: Building a foundation for gene family analysis in Rosaceae genomes with a novel work flow: a case study in Pyrus architecture genes. Front. Plant Sci. 13, 975942 (2022)
Zheng, X., et al.: MdWRKY9 overexpression confers intensive dwarfing in the M26 rootstock of apple by directly inhibiting brassinosteroid synthetase MdDWF4 expression. New Phytol. 217(3), 1086–1098 (2018)
Hollender, C.A., Hadiarto, T., Srinivasan, C., Scorza, R., Dardick, C.: A brachytic dwarfism trait (dw) in peach trees is caused by a nonsense mutation within the gibberellic acid receptor PpeGID1c. New Phytol. 210(1), 227–239 (2016)
Jia, D., Gong, X., Li, M., Li, C., Sun, T., Ma, F.: Overexpression of a novel apple NAC transcription factor gene, MdNAC1, confers the Dwarf phenotype in transgenic apple (Malus domestica). Genes (Basel). 9(5), 1–18 (2018b)
Johnson, J.M., Kohler, A.R., Haus, M.J., Hollender, C.A.: Arabidopsis weep mutants exhibit narrow root angles. Micro Publ. Biol. 2022 (2022). https://doi.org/10.17912/micropub.biology.000584
Waite, J.M., Dardick, C.: The roles of the IGT gene family in plant architecture: past, present, and future. Curr. Opin. Plant Biol. 59, 101983 (2021)
Hollender, C.A., et al.: Loss of a highly conserved sterile alpha motif domain gene (WEEP) results in pendulous branch growth in peach trees. Proc. Natl. Acad. Sci. U. S. A. 115(20), E4690–E4699 (2018)
Hollender, C.A., Hill, J.L., Waite, J., Dardick, C.: Opposing influences of TAC1 and LAZY1 on lateral shoot orientation in Arabidopsis. Sci. Rep. 10(1), 1–13 (2020)
Taniguchi, M., et al.: The arabidopsis LAZY1 family plays a key role in gravity signaling within statocytes and in branch angle control of roots and shoots. Plant Cell. 29(8), 1984–1999 (2017)
Salojärvi, J., et al.: Genome sequencing and population genomic analyses provide insights into the adaptive landscape of silver birch. Nat. Genet. 49(6), 904–912 (2017)
Petersen, R., Krost, C.: Tracing a key player in the regulation of plant architecture: the columnar growth habit of apple trees (Malus × domestica). Planta. 238(1), 1–22 (2013)
Otto, D., Petersen, R., Schmidt, E.R., Gypsy, Á.T., Leaf, I.Á.R.Á.: The columnar mutation (“‘Co gene’”) of apple (Malus x domestica) is associated with an integration of a Gypsy-like retrotransposon. Mol. Breed. 33, 863–880 (2014)
Dougherty, L., Bai, T., Brown, S., Xu, K., Silva, H.: Exploring DNA variant segregation types enables mapping loci for recessive phenotypic suppression of Columnar growth in apple. Front. Plant Sci. 11, 692 (2020)
Han, T., Yu, J., Zhuang, J., Wang, Z., Sun, X., Zhang, Y.: The characterization of columnar apple gene MdCoL promoter and its response to abscisic acid, brassinosteroid and gibberellic acid. Int. J. Mol. Sci. 23(18), 1–15 (2022)
Flachowsky, H., Hättasch, C., Höfer, M., Peil, A., Hanke, M.V.: Overexpression of LEAFY in apple leads to a columnar phenotype with shorter internodes. Planta. 231(2), 251–263 (2010)
Sassa, H., Hirano, H., Ikehashi, H.: Self-incompatibility-related RNases in styles of Japanese pear (Pyrus serotina Rehd.). Plant Cell Physiol. 33(6), 811–814 (1992)
Okada, K., et al.: Related polymorphic F-box protein genes between haplotypes clustering in the BAC contig sequences around the S-RNase of Japanese pear. J. Exp. Bot. 62(6), 1887–1902 (2011)
DeNettancourt, D.: Incompatibility in angiosperms. Sex. Plant Reprod. 10(4), 185–199 (1997)
Sassa, H., Hirano, H., Nishio, T., Koba, T.: Style-specific self-compatible mutation caused by deletion of the S-RNase gene in Japanese pear (Pyrus serotina). Plant J. 12(1), 223–227 (1997)
Sassa, H., et al.: S locus F-box brothers: multiple and pollen-specific F-box genes with S haplotype-specific polymorphisms in apple and Japanese pear. Genetics. 175(4), 1869–1881 (2007)
Okada, K., et al.: Deletion of a 236 kb region around S4-RNase in a stylar-part mutant S4 sm-haplotype of Japanese pear. Plant Mol. Biol. 66(4), 389–400 (2008)
Kubo, K., et al.: Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science. 330(6005), 796–799 (2010)
De Franceschi, P., Dondini, L., Sanzol, J.: Molecular bases and evolutionary dynamics of self-incompatibility in the Pyrinae (Rosaceae). J. Exp. Bot. 63(11), 4015–4032 (2012)
Broothaerts, W., Keulemans, J., Van Nerum, I.: Self-fertile apple resulting from S-RNase gene silencing. Plant Cell Rep. 22(7), 497–501 (2004)
Li, M.F., Li, X.F., Han, Z.H., Shu, H.R., Li, T.Z.: Molecular analysis of two Chinese pear (Pyrus bretschneideri Rehd.) spontaneous self-compatible mutants, Yan Zhuang and Jin Zhui. Plant Biol. 11(5), 774–783 (2009a)
Sanzol, J.: Pistil-function breakdown in a new S-allele of European pear, S21°, confers self-compatibility. Plant Cell Rep. 28(3), 457–467 (2009)
Sawamura, Y., et al.: A self-compatible Pollen-part mutant of Japanese pear produced by crossing “Kosui” with Pollen from Gamma-irradiated “Kosui”. J. Japanese Soc. Hortic. Sci. 82(3), 222–226 (2013)
Mase, N., et al.: A segmental duplication encompassing S-haplotype triggers pollen-part self-compatibility in Japanese pear (Pyrus pyrifolia). Mol. Breed. 33(1), 117–128 (2014)
Gustafson, F.G.: Parthenocarpy: natural and artificial. Bot. Rev. 8(9), 599–654 (1942)
Moniruzzaman, M., Darwish, A.G., Ismail, A., El-kereamy, A., Tsolova, V., El-Sharkawy, I.: Seedlessness trait and genome editing—a review. Int. J. Mol. Sci. 24(6), 5660 (2023)
Joldersma, D., Liu, Z.: The making of virgin fruit: the molecular and genetic basis of parthenocarpy. J. Exp. Bot. 69(5), 955–962 (2018)
Nishitani, C., et al.: Parthenocarpic genetic resources and gene expression related to parthenocarpy among four species in pear (Pyrus spp.). Sci. Hortic. (Amsterdam). 136, 101–109 (2012)
Quinet, M., Buyens, C., Dobrev, P.I., Motyka, V., Jacquemart, A.L.: Hormonal regulation of early fruit development in European pear (Pyrus communis cv. “conference”). Horticulturae. 5(1), 1–18 (2019)
Griggs, W.H., Iwakiri, B.T.: Pollination and parthenocarpy in the production of Bartlett pears in California. Hilgardia. 22(19), 643–678 (1954)
Pauwels, E., Eyssen, R., Keulemans, J.: Parthenocarpy and apple breeding. Acta Hortic. 484, 55–59 (1999)
Lordan, J., Vilardell, P., Peris, M., Torres, E., Alegre, S., Asín, L.: Post petal fall applications of gibberellins improve fruit set on pear. Sci. Hortic. (Amsterdam). 252, 149–155 (2019)
Cong, L., et al.: CPPU may induce gibberellin-independent parthenocarpy associated with PbRR9 in “Dangshansu” pear. Hortic. Res. 7(1), 1–14 (2020)
Ren, Z., Li, Z., Miao, Q., Yang, Y., Deng, W., Hao, Y.: The auxin receptor homologue in Solanum lycopersicum stimulates tomato fruit set and leaf morphogenesis. J. Exp. Bot. 62(8), 2815–2826 (2011)
Mazzucato, A., et al.: A TILLING allele of the tomato Aux/IAA9 gene offers new insights into fruit set mechanisms and perspectives for breeding seedless tomatoes. Mol. Breed. 35(22), 15 (2015)
Goetz, M., Hooper, L.C., Johnson, S.D., Rodrigues, J.C.M., Vivian-Smith, A., Koltunow, A.M.: Expression of aberrant forms of AUXIN RESPONSE FACTOR8 stimulates parthenocarpy in arabidopsis and tomato. Plant Physiol. 145(2), 351–366 (2007)
Molesini, B., Pandolfini, T., Rotino, G.L., Dani, V., Spena, A.: Aucsia gene silencing causes parthenocarpic fruit development in tomato. Plant Physiol. 149(1), 534–548 (2009)
García-Hurtado, N., et al.: The characterization of transgenic tomato overexpressing gibberellin 20-oxidase reveals induction of parthenocarpic fruit growth, higher yield, and alteration of the gibberellin biosynthetic pathway. J. Exp. Bot. 63(16), 5803–5813 (2012)
Martínez-Bello, L., Moritz, T., López-Díaz, I.: Silencing C 19 -GA 2-oxidases induces parthenocarpic development and inhibits lateral branching in tomato plants. J. Exp. Bot. 66(19), 5897–5910 (2015)
Martí, C., Orzáez, D., Ellul, P., Moreno, V., Carbonell, J., Granell, A.: Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J. 52(5), 865–876 (2007)
Mazzucato, A., Taddei, A.R., Soressi, G.P.: The parthenocarpic fruit (pat) mutant of tomato (Lycopersicon esculentum Mill.) sets seedless fruits and has aberrant anther and ovule development. Development. 125(1), 107–114 (1998)
Fuentes, S., Ljung, K., Sorefan, K., Alvey, E., Harber, N.P., Østergaard, L.: Fruit growth in Arabidopsis occurs via DELLA-dependent and DELLA-independent gibberellin responses. Plant Cell. 24(10), 3982–3996 (2012)
Mazzucato, A., Olimpieri, I., Siligato, F., Picarella, M.E., Soressi, G.P.: Characterization of genes controlling stamen identity and development in a parthenocarpic tomato mutant indicates a role for the DEFICIENS ortholog in the control of fruit set. Physiol. Plant. 132(4), 526–537 (2008)
Medina, M., et al.: Early anther ablation triggers parthenocarpic fruit development in tomato. Plant Biotechnol. J. 11(6), 770–779 (2013)
De Martino, G., Pan, I., Emmanuel, E., Levy, A., Irish, V.F.: Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell. 18(8), 1833–1845 (2006)
Niu, S., He, Y., Yan, S., Sun, Z., Cai, R., Zhang, Y.: Histological, transcriptomic, and gene functional analyses reveal the regulatory events underlying gibberellin-induced parthenocarpy in tomato. Hortic. Plant J. 12, 1–16 (2023)
Okabe, Y., et al.: Aberrant stamen development is associated with Parthenocarpic fruit set through up-regulation of gibberellin biosynthesis in tomato. Plant Cell Physiol. 60(1), 38–51 (2019)
Tanaka, N., Wada, M.: Apple MADS genes are involved in Parthenocarpy and floral organ formation. Hortic. J. 91(2), 131–139 (2022)
Yao, J.-L.: Parthenocarpic apple fruit production conferred by transposon insertion mutations in a MADS-box transcription factor. Proc. Natl. Acad. Sci. 98(3), 1306–1311 (2001)
Cong, L., et al.: 2,4-D-induced parthenocarpy in pear is mediated by enhancement of GA 4 biosynthesis. Physiol. Plant. 166(3), 812–820 (2019)
Liu, L., et al.: Histological, hormonal and transcriptomic reveal the changes upon gibberellin-induced parthenocarpy in pear fruit. Hortic. Res. 5(1), 1–13 (2018)
Flachowsky, H., Hanke, M.V., Peil, A., Strauss, S.H., Fladung, M.: A review on transgenic approaches to accelerate breeding of woody plants: review. Plant Breed. 128(3), 217–226 (2009)
Lang, G.A., Early, J.D., Martin, G.C., Darnell, R.L.: Endo-, Para-, and Ecodormancy: physiological terminology and classification for dormancy research. HortScience. 22(3), 1–20 (1987)
Faust, M., Erez, A., Rowland, L.J.J., Wang, S.Y.Y., Norman, H.A.A.: Bud dormancy in perennial fruit trees. HortScience. 32(4), 623–629 (1997)
Howe, G.T., et al.: Physiological and genetic approaches to studying endodormancy-related traits in Populus. HortScience. 34(7), 1174–1184 (1999)
Yang, Q., Gao, Y., Wu, X., Moriguchi, T., Bai, S., Teng, Y.: Bud endodormancy in deciduous fruit trees: advances and prospects. Hortic. Res. 8(1), 139 (2021)
Tominaga, A., Ito, A., Sugiura, T., Yamane, H.: How is global warming affecting fruit tree blooming? “Flowering (dormancy) disorder” in Japanese pear (Pyrus pyrifolia) as a case study. Front. Plant Sci. 12(2), 1–17 (2022)
El-Yazal, M.A.S., Rady, M.M., Seif, S.A.: Foliar-applied dormancy-breaking chemicals change the content of nitrogenous compounds in the buds of apple (Malus sylvestris Mill. cv.Anna) trees. J. Hortic. Sci. Biotechnol. 87(4), 299–304 (2012)
Li, J., Xu, Y., Niu, Q., He, L., Teng, Y., Bai, S.: Abscisic acid (ABA) promotes the induction and maintenance of pear (Pyrus pyrifolia white pear group) flower bud endodormancy. Int. J. Mol. Sci. 19(1), 1–20 (2018)
Yang, Q., et al.: PpyGAST1 is potentially involved in bud dormancy release by integrating the GA biosynthesis and ABA signaling in “Suli” pear (Pyrus pyrifolia White Pear Group). Environ. Exp. Bot. 162, 302–312 (2019)
Tuan, P.A., Bai, S., Saito, T., Ito, A., Moriguchi, T.: Dormancy-associated MADS-box (DAM) and the abscisic acid pathway regulate pear endodormancy through a feedback mechanism. Plant Cell Physiol. 58(8), 1378–1390 (2017)
Yang, Q., et al.: ABA-responsive ABRE-BINDING FACTOR3 activates DAM3 expression to promote bud dormancy in Asian pear. Plant Cell Environ. 43(6), 1360–1375 (2020)
Kumar, G., Rattan, U.K., Singh, A.K.: Chilling-mediated DNA methylation changes during dormancy and its release reveal the importance of epigenetic regulation during winter dormancy in Apple (Malus x domestica Borkh.). PLoS One. 11(2), 1–25 (2016)
Saito, T., Bai, S., Imai, T., Ito, A., Nakajima, I., Moriguchi, T.: Histone modification and signalling cascade of the dormancy-associatedMADS-box gene, PpMADS13-1, in Japanese pear (Pyrus pyrifolia) during endodormancy. Plant Cell Environ. 38(6), 1157–1166 (2015)
Li, Z., Reighard, G.L., Abbott, A.G., Bielenberg, D.G.: Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J. Exp. Bot. 60(12), 3521–3530 (2009b)
Salama, A.M., et al.: Temperate fruit trees under climate change: challenges for dormancy and chilling requirements in warm winter regions. Horticulturae. 7(86), 1–14 (2021)
Drepper, B., Gobin, A., Van Orshoven, J.: Spatio-temporal assessment of frost risks during the flowering of pear trees in Belgium for 1971–2068. Agric. For. Meteorol. 315, 108822 (2022)
Hanke, M.-V., Flachowsky, H., Peil, A., Hättasch, C.: No flower no fruit – genetic potentials to trigger flowering in fruit trees. Genes, Genomes and Genomics. 1(1), 1–20 (2007)
Jackson, D., Looney, N., Morley-Bunker, M.: Temperate and Subtropical Fruit Production, pp. 327. CAB International, Oxford UK, ISBN: 978-1-84593-501-6 (2010)
Washington State University: Apple Chemical Thinning | WSU Production Guide. WSU (2023)
Turner, J.N.: Gibberellic acid for controlling fruit production of pears. Acta Hortic. 34, 287–298 (1972)
Sharma, N., Singh, S.K., Mahato, A.K., Ravishankar, H., Dubey, A.K., Singh, N.K.: Physiological and molecular basis of alternate bearing in perennial fruit crops. Sci. Hortic. (Amsterdam). 243, 214–225 (2019)
Callejas, R., Bangerth, F.: Is auxin export of apple fruit an alternative signal for inhibition of flower bud induction? Acta Hortic. 463, 271–277 (1998)
Milyaev, A., Tandron-Moya, Y.A., von Wirén, N., Neuwald, D., Flachowsky, H., Wünsche, J.N.: What else don’t we know about biennial bearing? Phytohormone profile of seeds and seed number per fruit differ between a biennial and a non-biennial apple cultivar. Acta Hortic. 1342, 7–14 (2022a)
Milyaev, A., et al.: Profiling of phytohormones in apple fruit and buds regarding their role as potential regulators of flower bud formation. Tree Physiol. 42(11), 2319–2335 (2022b)
Guitton, B., Kelner, J.J., Velasco, R., Gardiner, S.E., Chagné, D., Costes, E.: Genetic control of biennial bearing in apple. J. Exp. Bot. 63(1), 131–149 (2012)
Wang, M., et al.: BRANCHED1: a key hub of shoot branching. Front. Plant Sci. 10(2), 1–12 (2019)
Guitton, B., et al.: Analysis of transcripts differentially expressed between fruited and deflowered “gala” adult trees: a contribution to biennial bearing understanding in apple. BMC Plant Biol. 16(1), 1–22 (2016)
Turner, J.N.: GA for controlling fruit production of pears. Acta Hortic. Symp. Growth Regul. Fruit Prod. 34, 287–297 (1973)
Fan, S., et al.: Dynamic cytosine DNA methylation patterns associated with mRNA and siRNA expression profiles in alternate bearing apple trees. J. Agric. Food Chem. 67(18), 5250–5264 (2019)
Singh, S., Chaudhary, R., Deshmukh, R., Tiwari, S.: Opportunities and challenges with CRISPR-Cas mediated homologous recombination based precise editing in plants and animals. Plant Mol. Biol. 111, 1–20 (2023)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2024 The Author(s)
About this chapter
Cite this chapter
Claessen, H., Aert, P., De Storme, N. (2024). Targeted Gene Editing in Pome Fruit Genetics and Breeding: State-of-the-Art, Application Potential and Perspectives. In: Ricroch, A., Eriksson, D., Miladinović, D., Sweet, J., Van Laere, K., Woźniak-Gientka, E. (eds) A Roadmap for Plant Genome Editing . Springer, Cham. https://doi.org/10.1007/978-3-031-46150-7_19
Download citation
DOI: https://doi.org/10.1007/978-3-031-46150-7_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-46149-1
Online ISBN: 978-3-031-46150-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)