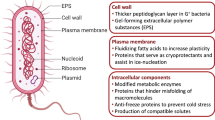
Abstract
Antarctica is covered with ice and therefore the coldest, driest and least populated continent of the Earth. Apart from the challenges of sustaining the life in such environment, the microbial diversity of archaea, bacteria and fungi can thrive here by maintaining the membrane fluidity, producing the antifreeze proteins, cold-shock proteins, cryoprotectants, osmolytes, antioxidants, cold active enzymes, alteration in DNA, etc. and thereby adapted for the cold environments. In Antarctica, these microbes are the main basis for the biogeocycling of nutrients in extreme environments. The enzymes produced by these psychrophilic organisms are cold active and therefore gained the spotlight owing to their significance in environment and research. The behaviour of these cold-active enzymes has a wide and potentially biotechnological applications in fields as wide as the detergent, textile and food industries, medical and pharmaceutical preparations, bioremediation, etc. Therefore, the current manuscript illustrates the native microbial diversity and their significance and application aspect in environment and industry.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Fretwell P, Pritchard HD, Vaughan DG, Bamber JL et al (2013) Bedmap 2: improved ice bed, surface and thickness datasets for Antarctica. Cryosphere 7:375–393
Cowan DA, Tow LA (2004) Endangered Antarctic environments. Annu Rev Microbiol 58:649–690. https://doi.org/10.1146/annurev.micro.57.030502.090811
Chong CW, Pearce DA, Convey P (2015) Emerging spatial patterns in Antarctic prokaryotes. Front Microbiol 6:1058. https://doi.org/10.3389/fmicb.2015.01058
Sadaiappan B, Kannan S, Palaniappan S, Manikkam R, Ramasamy B, Anilkumar N, Subramanian M (2020) Metagenomic 16S rDNA amplicon data of microbial diversity and its predicted metabolic functions in the Southern Ocean (Antarctic). Data Br 28:104876
Cavicchioli R, Charlton T, Ertan H, Mohd Omar S, Siddiqui KS, Williams TJ (2011) Biotechnological uses of enzymes from psychrophiles. J Microbial Biotech 4:449
Bottos EM, Laughlin DC, Herbold CW, Lee CK, McDonald IR, Cary SC (2020) Abiotic factors influence patterns of bacterial diversity and community composition in the Dry Valleys of Antarctica. FEMS Microbiol Ecol 96(5):fiaa042
Magalhaes C, Stevens MI, Cary SC, Ball BA, Storey BC, Wall DH, Ruprecht U (2012) At limits of life: multidisciplinary insights reveal environmental constraints on biotic diversity in continental Antarctica. PLos One 7(9):e44578. https://doi.org/10.1371/journal.pone.0044578
Velasco-Castrillon A, Schultz MB, Colombo F, Gibson JA, Davies KA, Austin AD, Stevens MI (2014) Distribution and diversity of soil microfauna from East Antarctica: assessing the link between biotic and abiotic factors. PLoS One 9(1):e87529
Convey P (1997) How are the life history strategies of Antarctic terrestrial invertebrates influenced by extreme environmental conditions? J Therm Biol 22(6):429–440
Aronson RB, Thatje S, McClintock JB, Hughes KA (2011) Anthropogenic impacts on marine ecosystems in Antarctica. Ann N Y Acad Sci 1223(1):82–107
Padeiro A, Amaro E, Dos Santos MM, Araújo MF, Gomes SS, Leppe M et al (2016) Trace element contamination and availability in the Fildes Peninsula, King George Island, Antarctica. Environ Sci Process Impacts 18(6):648–657
Ogaki MB, Coelho LC, Vieira R, Neto AA, Zani CL, Alves TM et al (2020) Cultivable fungi present in deep-sea sediments of Antarctica: taxonomy, diversity, and bioprospecting of bioactive compounds. Extremophiles 24:227–238
Rosa Luiz H et al (2019) Fungi in Antarctica: diversity, ecology, effects of climate change, and bioprospection for bioactive compounds. In: Fungi of Antarctica. Springer, Cham, pp 1–17
Tedersoo L, Sánchez-Ramírez S, Köljalg U, Bahram M, Döring M, Schigel D, May T, Ryberg M, Abarenkov K (2018) High-level classification of the fungi and a tool for evolutionary ecological analyses. Fungal Div 90:135–159
Bridge PD, Spooner BM (2012) Non-lichenized Antarctic fungi: transient visitors or members of a cryptic ecosystem? Fungal Ecol 5:381–394
Ruisi S, Barreca D, Selbmann L, Zucconi L, Onofri S (2007) Fungi in Antarctica. Rev Environ Sci Biotechnol 6:127–141
Cary SC, McDonald IR, Barrett JE, Cowan DA (2010) On the rocks: the microbiology of Antarctic Dry Valley soils. Nat Rev Microbiol 8:129–138
Núñez-Montero K, Barrientos L (2018) Advances in Antarctic research for antimicrobial discovery: A comprehensive narrative review of bacteria from Antarctic environments as potential sources of novel antibiotic compounds against human pathogens and microorganisms of industrial importance. Antibiotics 7:90. https://doi.org/10.3390/antibiotics7040090
Teixeira LCRS, Peixoto RS, Cury JC, Sul WJ, Pellizari VH, Tiedje J, Rosado AS (2010) Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. ISME J 4:989
O’Brien A, Sharp R, Russell NJ, Roller S (2004) Antarctic bacteria inhibit growth of food-borne microorganisms at low temperatures. FEMS Microbiol Ecol 48:157–167
Hultman J, Waldrop M, Mackelprang R et al (2015) Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes. Nature 521:208–212. https://doi.org/10.1038/nature14238
Dhaulaniya AS, Balan B, Agrawal PK, Singh DK (2019) Cold survival strategies for bacteria, recent advancement and potential industrial applications. Arch Microbiol 201(1):1–16
Ghobakhlou AF, Johnston A, Harris L, Antoun H, Laberge S (2015) Microarray transcriptional profiling of Arctic Mesorhizobium strain N33 at low temperature provides insights into cold adaption strategies. BMC Genomics 16(1):1–14
Nagy G, Kerekes R (1981) Influence of cultivation temperature on the TTC-reducing capacity of psychrophilic, psychotropic, and mesophilic pseudomonas strains. ZentralblattfürBakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene. ZweiteNaturwissenschaftlicheAbteilung: Mikrobiologie der Landwirtschaft, der Technologie und des Umweltschutzes 136(3):185–188
Ayala-del-Río HL, Chain PS, Grzymski JJ, Ponder MA, Ivanova N, Bergholz PW et al (2010) The genome sequence of Psychrobacter arcticus 273-4, a psychoactive Siberian permafrost bacterium, reveals mechanisms for adaptation to low-temperature growth. Appl Environ Microbiol 76(7):2304–2312
Celik Y, Graham LA, Mok YF, Bar M, Davies PL, Braslavsky I (2010) Superheating of ice crystals in antifreeze protein solutions. Proc Natl Acad Sci 107(12):5423–5428
Muñoz PA, Márquez SL, González-Nilo FD, Márquez-Miranda V, Blamey JM (2017) Structure and application of antifreeze proteins from Antarctic bacteria. Microb Cell Factories 16(1):1–13
Woldringh CL, Jensen PR, Westerhoff HV (1995) Structure and partitioning of bacterial DNA: determined by a balance of compaction and expansion forces? FEMS Microbiol Lett 131(3):235–242
Shivaji S, Prakash JS (2010) How do bacteria sense and respond to low temperature? Arch Microbiol 192(2):85–95
Halliwell B (2006) Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol 141(2):312–322
Correa-Llantén DN, Amenábar MJ, Blamey JM (2012) Antioxidant capacity of novel pigments from an Antarctic bacterium. J Microbiol 50(3):374–379
Khan A, Singh AV, Pareek N, Arya P, Upadhayay VK, Kumar Jugran A, Kumar Mishra P, Goel R (2023a) Credibility assessment of cold adaptive Pseudomonas jesenni MP1 and P. palleroniana N26 on growth, rhizosphere dynamics, nutrient status, and yield of the kidney bean cultivated in Indian Central Himalaya. Front Plant Sci 14:1042053. https://doi.org/10.3389/fpls.2023.1042053
Khan A, Panthari D, Sharma RS, Punetha A, Singh AV, Upadhayay VK (2023b) Biofertilizers: a microbial-assisted strategy to improve plant growth and soil health. In: Advanced microbial techniques in agriculture, environment, and health management. Academic Press, pp 97–118
Bardgett RD, Van Der Putten WH (2014) Belowground biodiversity and ecosystem functioning. Nature 515(7528):505–511
Khan A, Singh AV (2021) Multifarious effect of ACC deaminase and EPS producing Pseudomonas sp. and Serratia marcescens to augment drought stress tolerance and nutrient status of wheat. World J Microbiol Biotechnol 37(12):1–17
Upadhayay VK, Singh AV, Khan A (2022a) Cross talk between zinc-solubilizing bacteria and plants: A short tale of bacterial-assisted zinc biofortification. Frontiers in Soil Science 1(788170):10–3389
Khan A, Upadhayay VK, Panwar M, Singh AV (2020a) Soil microbiota: A key bioagent for revitalization of soil health in hilly regions. In: Microbiological advancements for higher Altitude Agro-Ecosystems & Sustainability. Springer, Singapore, pp 183–200
de la Torre JR, Goebel BM, Friedmann EI, Pace NR (2003) Microbial diversity of cryptoendolithic communities from the McMurdo Dry Valleys, Antarctica. Appl Environ Microbiol 69(7):3858–3867
Straza TR, Cottrell MT, Ducklow HW, Kirchman DL (2009) Geographic and phylogenetic variation in bacterial biovolume as revealed by protein and nucleic acid staining. Appl Environ Microbiol 75(12):4028–4034
Badger MR, Bek EJ (2008) Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J Exp Bot 59(7):1525–1541
Allan J, Ronholm J, Mykytczuk NCS, Greer CW, Onstott TC, Whyte LG (2014) Methanogen community composition and rates of methane consumption in Canadian high Arctic permafrost soils. Environ Microbiol Rep 6(2):136–144
Tveit AT, Urich T, Frenzel P, Svenning MM (2015) Metabolic and trophic interactions modulate methane production by Arctic peat microbiota in response to warming. Proc Natl Acad Sci 112(19):E2507–E2516
Khan A, Joshi M, Singh AV (2020b) Rhizospheric microbial community: ecology, methods, and functions. In: Sharma SK, Singh UB, Sahu PK, Singh HV, Sharma PK (eds) Rhizosphere microbiology. Springer, Singapore, pp 127–148
Ortiz M, Bosch J, Coclet C, Johnson J, Lebre P, Salawu-Rotimi A et al (2020) Microbial nitrogen cycling in Antarctic soils. Microorganisms 8(9):1442
Martínez-Pérez C, Mohr W, Löscher CR, Dekaezemacker J, Littmann S, Yilmaz P et al (2016) The small unicellular diazotrophic symbiont, UCYN-A, is a key player in the marine nitrogen cycle. Nat Microbiol 1(11):1–7
Vishniac HS (2006) Yeast biodiversity in the Antarctic. In: Biodiversity and ecophysiology of yeasts. Springer, Berlin, Heidelberg, pp 419–440
Chan Y, Van Nostrand JD, Zhou J, Pointing SB, Farrell RL (2013) Functional ecology of an Antarctic dry valley. Proc Natl Acad Sci 110(22):8990–8995
Singh J, Singh AV, Upadhayay VK, Khan A, Chandra R (2022) Prolific contribution of Pseudomonas protegens in Zn biofortification of wheat by modulating multifaceted physiological response under saline and non-saline conditions. World J Microbiol Biotechnol 38(12):1–20
Khan A, Singh AV, Upadhayay VK, Ballabh A, Prasad B (2022a) Influence of PGPR on growth and yield of oat (Avena sativa L.) under field conditions. Indian. J Ecol 49(4):1351–1356
Upadhayay VK, Singh AV, Khan A, Singh J, Pareek N, Raghav A (2022b) FE-SEM/EDX based zinc mobilization analysis of Burkholderia cepacia and Pantoea rodasii and their functional annotation in crop productivity, soil quality, and zinc biofortification of Paddy. Front Microbiol 13. https://doi.org/10.3389/fmicb.2022.852192
Khan A, Singh AV, Kumar R, Kukreti B, Bundela V, Upadhayay VK (2022b) Relative impact of PGPR inoculation on biofortification and yield of wheat under field conditions and their performance assessment through statistical tools. J Pharm Innov 11(8):490–495
Kaviya N, Upadhayay VK, Singh J, Khan A, Panwar M, Singh AV (2019) Role of microorganisms in soil genesis and functions. In: Mycorrhizosphere and Pedogenesis. Springer, Singapore, pp 25–52
Khan A, Singh J, Upadhayay VK, Singh AV, Shah S (2019) Microbial biofortification: A green technology through plant growth promoting microorganisms. In: Sustainable green technologies for environmental management. Springer, Singapore, pp 255–269
Punetha A, Punetha S, Khan A (2022) Soil community composition and ecosystem processes. In: Rukhsana, Alam A (eds) Agriculture, environment and sustainable development. Springer, Cham, pp 217–236
Upadhayay VK, Singh J, Khan A, Lohani S, Singh AV (2019) Mycorrhizal mediated micronutrients transportation in food based plants: A biofortification strategy. In: Mycorrhizosphere and Pedogenesis. Springer, Singapore, pp 1–24
Duarte AWF, Dos Santos JA, Vianna MV, Vieira JMF, Mallagutti VH, Inforsato FJ et al (2018) Cold-adapted enzymes produced by fungi from terrestrial and marine Antarctic environments. Crit Rev Biotechnol 38(4):600–619
Upadhayay VK, Khan A, Singh J, Singh AV (2021a) Bacterial assisted improved Zn consignment in root and shoot of rice plant by zinc solubilizing Serratia marcescens bearing plant probiotic traits. Adv Biores 13(1):1–8
Upadhayay VK, Singh AV, Khan A, Pareek N (2021b) Influence of zinc solubilizing bacterial co-inoculation with zinc oxide supplement on rice plant growth and Zn uptake. J Pharm Innov 10:113–116
Jorquera MA, Shaharoona B, Nadeem SM, de la Luz Mora M, Crowley DE (2012) Plant growth-promoting rhizobacteria associated with ancient clones of creosote bush (Larrea tridentata). Microb Ecol 64(4):1008–1017
Roshani KA, Singh AV, Upadhayay VK, Prasad B (2020) Development of potential microbial consortia and their assessment on wheat (Triticum aestivum) seed germination. Environ Ecol 38(1):6–16
Parveen H, Singh AV, Khan A, Prasad B, Pareek N (2018) Influence of plant growth promoting rhizobacteria on seed germination and seedling vigor of green gram. Int J Chem Stud 6(4):611–618
Fardella C, Oses R, Torres-Díaz C, Molina-Montenegro MA (2014) Antarctic fungal endophytes as tool for the reintroduction of native plant species in arid zones. Bosque 35:235–239. https://doi.org/10.4067/S0717-92002014000200011
Berríos G, Cabrera G, Gidekel M, Gutiérrez-Moraga A (2013) Characterization of a novel antarctic plant growth-promoting bacterial strain and its interaction with antarctic hair grass (Deschampsia Antarctica Desv). Polar Biol 36(3):349–362
Tomova I, Stoilova-Disheva M, Lazarkevich I, Vasileva-Tonkova E (2015) Antimicrobial activity and resistance to heavy metals and antibiotics of heterotrophic bacteria isolated from sediment and soil samples collected from two Antarctic islands. Front Life Sci 8(4):348–357
Presta L, Inzucchi I, Bosi E, Fondi M, Perrin E, Miceli E et al (2016) Draft genome sequence of Flavobacterium sp. strain TAB 87, able to inhibit the growth of cystic fibrosis bacterial pathogens belonging to the Burkholderia cepacia complex. Genome Announc 4(3):e00410–e00416
Khan A, Kukreti B, Makarana G, Suyal DC, Singh AV, Kumar S (2023c) Role of microorganisms in alleviation of arsenic toxicity in plants. In: Unravelling plant-microbe synergy. Academic Press, pp 263–281
Khan A, Sharma RS, Panthari D, Kukreti B, Singh AV, Upadhayay VK (2023d) Bioremediation of heavy metals by soil dwelling microbes: an environment survival approach. In: Pandey SC, Pande V, Sati D, Samant M (eds) Advanced microbial techniques in agriculture, environment and health management. Elsevier, London, pp 167–190
Ismail W, Gescher J (2012) Epoxy coenzyme A thioester pathways for degradation of aromatic compounds. Appl Environ Microbiol 78(15):5043–5051
Dias RL, Ruberto L, Hernández E, Vázquez SC, Balbo AL, Del Panno MT, Mac Cormack WP (2012) Bioremediation of an aged diesel oil-contaminated Antarctic soil: evaluation of the “on site” biostimulation strategy using different nutrient sources. Int Biodeterior Biodegradation 75:96–103
Pérez-de-Mora A, Engel M, Schloter M (2011) Abundance and diversity of n-alkane-degrading bacteria in a forest soil co-contaminated with hydrocarbons and metals: a molecular study on alkB homologous genes. Microb Ecol 62:959–972
Powell SM, Ferguson SH, Bowman JP, Snape I (2006) Using real-time PCR to assess changes in the hydrocarbon-degrading microbial community in Antarctic soil during bioremediation. Microb Ecol 52:523–532
Reyes-César A, Absalon AE, Fernández FJ, González JM, Cortés-Espinosa DV (2014) Biodegradation of a mixture of PAHs by non-ligninolytic fungal strains isolated from crude oil-contaminated soil. World J Microbiol Biotechnol 30:999–1009
Pini F, Grossi C, Nereo S, Michaud L, Giudice AL, Bruni V et al (2007) Molecular and physiological characterisation of psychotropic hydrocarbon-degrading bacteria isolated from Terra Nova Bay (Antarctica). Eur J Soil Biol 43(5–6):368–379
Aislabie JM, Balks MR, Foght JM, Waterhouse EJ (2004) Hydrocarbon spills on Antarctic soils: effects and management. Environ Sci Technol 38(5):1265–1274
Ruberto LA, Vazquez S, Lobalbo A, Mac Cormack WP (2005) Psychrotolerant hydrocarbon-degrading Rhodococcus strains isolated from polluted Antarctic soils. Antarct Sci 17(1):47–56
Stallwood B, Shears J, Williams PA, Hughes KA (2005) Low temperature bioremediation of oil-contaminated soil using biostimulation and bioaugmentation with a Pseudomonas sp. from maritime Antarctica. J Appl Microbiol 99(4):794–802
Saul DJ, Aislabie JM, Brown CE, Harris L, Foght JM (2005) Hydrocarbon contamination changes the bacterial diversity of soil from around Scott Base, Antarctica. FEMS Microbiol Ecol 53(1):141–155
Ma Y, Wang L, Shao Z (2006) Pseudomonas, the dominant polycyclic aromatic hydrocarbon-degrading bacteria isolated from Antarctic soils and the role of large plasmids in horizontal gene transfer. Environ Microbiol 8(3):455–465
Ferrari BC, Zhang C, Van Dorst J (2011) Recovering greater fungal diversity from pristine and diesel fuel contaminated sub-Antarctic soil through cultivation using both a high and a low nutrient media approach. Front Microbiol 2:217
Hua Z, Chen J, Lun S, Wang X (2003) Influence of biosurfactants produced by Candida Antarctica on surface properties of microorganism and biodegradation of n-alkanes. Water Res 37(17):4143–4150
Martorell MM, Ruberto LAM, Fernández PM, Castellanos de Figueroa LI, Mac Cormack WP (2017) Bioprospection of cold-adapted yeasts with biotechnological potential from Antarctica. J Basic Microbiol 57(6):504–516
Hughes KA, Bridge P, Clark MS (2007) Tolerance of Antarctic soil fungi to hydrocarbons. Sci Total Environ 372(2–3):539–548
Confalonieri F, Sommer S (2011) Bacterial and archaeal resistance to ionizing radiation. J Phys Conf Ser 261(1):012005. IOP Publishing
Das N, Chandran P (2011) Microbial degradation of petroleum hydrocarbon contaminants: an overview. Biotechnol Res Int 2011:941810. https://doi.org/10.4061/2011/941810
Rogan-Finnemore M (2005) The legal implications of bioprospecting in the Antarctic region. UC Research Repository
Zucconi L, Canini F, Temporiti ME, Tosi S (2020) Extracellular enzymes and bioactive compounds from Antarctic terrestrial fungi for bioprospecting. Int J Environ Res Public Health 17(18):6459
Zainal N, Ser HL, Yin WF, Tee KK, Lee LH, Chan KG (2016) Streptomyces humi sp. nov., an actinobacterium isolated from soil of a mangrove forest. Antonie Van Leeuwenhoek 109:467–474
Vercesi A, Nasini G, Locci R (1992) Biological and chemical characterization of the antibiotic activity of Streptomyces species isolated from grapevine carposphere. Actinomycetes 3(1)
Rojas JL, Martín J, Tormo JR, Vicente F, Brunati M, Ciciliato I et al (2009) Bacterial diversity from benthic mats of Antarctic lakes as a source of new bioactive metabolites. Mar Genomics 2(1):33–41
Teoh CP, Koh SP, Ling CMWV (2020) Characterisation of an Antarctic yeast, Glaciozyma Antarctica PI12. Borneo Int J Biotechnol 1:89
Fenice M (2016) The Psychrotolerant Antarctic fungus Lecanicillium muscarium CCFEE 5003: A powerful producer of cold-tolerant Chitinolytic enzymes. Molecules 21:447
Martorell MM, Ruberto LAM, de Figueroa LIC, Mac Cormack WP (2019) Antarctic Yeasts as a Source of Enzymes for Biotechnological Applications. In: Rosa LH (ed) Fungi of Antarctica: diversity, ecology and biotechnological applications. Springer International Publishing, Cham, pp 285–304
Baeza M, Alcaíno J, Cifuentes V, Turchetti B, Buzzini P (2017) Cold-active enzymes from cold-adapted yeasts. In: Sibirny AA (ed) Biotechnology of yeasts and filamentous fungi. Springer International Publishing, Cham, pp 297–324
He L, Mao Y, Zhang L, Wang H, Alias SA, Gao B (2017) Functional expression of a novel α-amylase from Antarctic psychrotolerant fungus for the baking industry and its magnetic immobilization. BMC Biotechnol 17:22
Mao Y, Yin Y, Zhang L, Alias SS, Gao B, Wei D (2015) Development of a novel Aspergillus uracil deficient expression system and its application in expressing a cold-adapted α-amylase gene from Antarctic fungi Geomyces pannorum. Process Biochem 50:1581–1590
Yu P, Wang XT, Liu JW (2015) Purification and characterization of a novel cold-adapted phytase from strain JMUY14 isolated from the Antarctic: characterization of a novel cold-adapted phytase. J Basic Microbiol 55:1029–1039
Białkowska AM, Szulczewska KM, Krysiak J, Florczak T, Gromek E, Kassassir H, Kur J, Turkiewicz M (2017) Genetic and biochemical characterization of yeasts isolated from Antarctic soil samples. Polar Biol 40:1787–1803
Salwoom L, Raja Abd Rahman RNZ, Salleh AB, Mohd Shariff F, Convey P, Pearce D, Mohamad Ali MS (2019) Isolation, characterisation, and lipase production of a cold-adapted bacterial strain Pseudomonas sp. LSK25 isolated from Signy Island, Antarctica. Molecules 24(4):715
Furbino LE, Pellizzari FM, Neto PC, Rosa CA, Rosa LH (2018) Isolation of fungi associated with macroalgae from maritime Antarctica and their production of agarolytic and carrageenolytic activities. Polar Biol 41:527–535
Marizcurrena JJ, Martı’nez-Lo’pez W, Ma H, Lamparter T, Castro-Sowinski S (2018) A highly efficient and cost-effective recombinant production of a bacterial photolyase from the Antarctic isolate Hymenobacter sp. UV11. Extremophiles 23:49
Gil-Dura’n C, Ravanal MC, Ubilla P, Vaca I, Cha’vez R (2018) Heterologous expression, purification and characterization of a highly thermolabile endoxylanase from the Antarctic fungus Cladosporium sp. Fungal Biol 122:87582
See-Too WS, Convey P, Pearce DA, Chan KG (2018) Characterization of a novel N-acyl homoserine lactonase, AidP, from Antarctic Planococcus sp. Microb Cell Factories 17:179
Dharmaraj S, Ashokkumar B, Dhevendaran K (2009) Fermentative production of carotenoids from marine actinomycetes. Iran J Microbiol 1(4):36–41
Namazkar S, Ahmad WA (2013) Spray-dried prodigiosin from Serratia marcescens as a colorant. Biosci Biotechnol Res Asia 10(1):69
Hoyoux A, Jennes I, Dubois P, Genicot S, Dubail F, François JM et al (2001) Cold-adapted β-galactosidase from the Antarctic psychrophile Pseudoalteromonas haloplanktis. Appl Environ Microbiol 67(4):1529–1535
Kavitha M (2016) Cold active lipases—an update. Front Life Sci 9:226–238
Araujo R, Casal M, Cavaco-Paulo A (2008) Application of enzymes for textile fibres processing. Biocatal Biotransformation 26:332–349. https://doi.org/10.1080/10242420802390457
Bisht S (2011) Cold active proteins in food and pharmaceutical industry [article on the Internet]. Biotech Articles. [cited 2017 Dec. 15]. Available from http://www.biotecharticles.com/Biotechnology-products-Article/ColdActive-Proteins-in-Food-and-Pharmaceutical-Industry-719
Hamid B, Bashir Z, Yatoo AM, Mohiddin F, Majeed N, Bansal M, Poczai P, Almalki WH, Sayyed RZ, Shati AA et al (2022) Cold-active enzymes and their potential industrial applications—a review. Molecules 27:5885. https://doi.org/10.3390/molecules27185885
Furhan J, Awasthi P, Sharma S (2019) Biochemical characterization and homology modeling of cold-active alkophilic protease from Northwestern Himalayas and its application in detergent industry. Biocatal Agric Biotechnol 17:726–735
Castilla A, Giordano SR, Irazoqui G (2022) Extremophilic lipases and esterases: characteristics and industrial applications. In: Kuddus M (ed) Microbial Extremozymes. Academic Press, pp 207–222
Ganasen M, Yaacob N, Rahman RN, Leow AT, Basri M, Salleh AB et al (2016) Cold-adapted organic solvent tolerant alkalophilic family I.3 lipase from an Antarctic pseudomonas. Int J Biol Macromol 92:1266–1276
Viñarta SC, Maza DD, Fernández PM, Aybar MJ, de Figueroa LIC (2022) Integrated production of biodiesel and industrial wastewater treatment by culturing oleaginous microorganisms. In: Kumar V, Kumar M (eds) Integrated environmental technologies for wastewater treatment and sustainable development. Elsevier, Amsterdam, pp 81–101
Plaza DO, Gallardo C, Straub YD, Bravo D, Pérez-Donoso JM (2016) Biological synthesis of fluorescent nanoparticles by cadmium and tellurite resistant Antarctic bacteria: exploring novel natural nanofactories. Microb Cell Factories 15(1):1–11
John MS, Nagoth JA, Ramasamy KP, Ballarini P, Mozzicafreddo M, Mancini A et al (2020) Horizontal gene transfer and silver nanoparticles production in a new Marinomonas strain isolated from the Antarctic psychrophilic ciliate Euplotes focardii. Sci Rep 10(1):1–14
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Khan, A. et al. (2023). Microbial Community Dynamics of Antarctica: Their Ecological Potential and Industrial Importance. In: Soni, R., Suyal, D.C., Morales-Oyervides, L. (eds) Microbial Bioactive Compounds. Springer, Cham. https://doi.org/10.1007/978-3-031-40082-7_15
Download citation
DOI: https://doi.org/10.1007/978-3-031-40082-7_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-40081-0
Online ISBN: 978-3-031-40082-7
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)