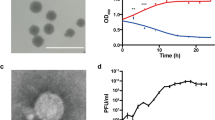
Abstract
The ability to rapidly adapt to changes in their environment is essential for the survival of microorganisms. DnaK is the prokaryotic analogue of eukaryotic Hsp70. These proteins protect the cell against heat injury or other forms of stress. In the present research, 78 rhizobial isolates were collected from different geographical regions of India and tested for their sensitivity. Forty-two out of 78 were shown to be susceptible to eight isolated phages. Only six Rhizobium strains out of 42 (MPSR033, MPSR041, MPSR052, MPSR084, MPSR220 and UPSR095) showed susceptibility to a single phage strain separately. All of these six rhizobial strains were slow growing and had the capability of Ex-planta nitrogenase activities. The PCR amplification of the dnaK region of the selected strain showed a single band ~ 600 bp in length. The restriction endonuclease analysis of dnaK regions produced six restriction patterns. Sequence BLASTn analysis of DnaK showed linkage with Actinobacterium sp. and Bradyrhizobium sp. Multiple sequence alignment of all six Rhizobium dnaK regions exhibited a 99 % similarity between MPSR041 and MPSR220, and formed a close cluster with UPSR095 and MPSR033. The Prosite database showed that the functional domain of the DnaK protein presented three signatures, PS00297 IDLGTTNS (HSP70_1), PS00329 VYDLGGGTFDISIL (HSP70_2) and PS01036 VVLVGGMSRMPKVQE (HSP70_3), which belong to the HSP70 protein family. An in silico interaction study revealed that DnaK protein is involved in interactions with blr, gap, grpE, pgk, hrcA and recA heat-shock proteins.







Similar content being viewed by others
References
Adams MH (1959) Bacteriophages. Wiley, New York
Appunu C, Dhar B (2008) Morphology and general characteristics of lytic phages infective on strains of Bradyrhizobium japonicum. Curr Microbiol 56:21–27
Boorstein WR, Zeigelhoffer T, Craig EA (1994) Molecular evolution of the HSP70 multigene family. J Mol Evol 38:1–17
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2(9):1511–1519
Craig EA, Kang PJ, Boorstein W (1990) A review of the role of 70 k Da heat shock proteins in protein translocation across membranes. Antonie von Leeuwenhoek 58:137–146
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Groemping Y, Reinstein J (2001) Folding properties of the nucleotide exchange factor GrpE from Thermus thermophilus: GrpE is a thermosensor that mediates heat shock response. J Mol Biol 314:167–178
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-Pdbviewer: an environment for comparative protein modeling. Electrophoresis 18(15):2714–23
Gupta RS, Singh B (1992) Cloning of the HSP70 gene from Halobacterium marismortui: relatedness of archaebacterial HSP70 to its eubacterial homologs and a model for the evolution of the HSP70 gene. J Bacteriol 174:4594–4605
Gupta RS, Singh B (1994) Phylogenetic analysis of 70 kDa heat shock protein sequences suggests a chimeric origin for the eukaryotic cell nucleus. Curr Biol 4:1104–1114
Hartl FU (1996) Molecular chaperones in cellular protein folding. Nature 381:571–580
Jaiswal SK, Dhar B (2010) Morphology and general characteristics of phages specific for Lens culinaris rhizobia. Biol Fertil Soils 46:681–687
Jaiswal SK, Dhar B (2011) Identification and assessment of symbiotic effectiveness of phage-typed Rhizobium leguminosarum strains on lentil (Lens culinaris Medik) cultivars. Curr Microbiol 62:1503–1509
Jaiswal SK, Dhar B, Rai AK (2012) Physiological and molecular characterization of locally adapted Rhizobium strains of lentil (Lens culinaris Medik) having restricted phage sensitivity. Ann Microbiol 62:1453–1459
Johnson C, Chandrasekhar GN, Georgopoulos C (1989) Escherechia coli DnaK and GrpE heat shock proteins interact both in vivo and in vitro. J Bacteriol 171:1590–1596
Lambert C, Léonard N, De Bolle X, Depiereux E (2002) ESyPred3D: prediction of proteins 3D structures. Bioinformatics 18(9):1250–6
Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8(4):477–486
Lindquist S (1986) The heat-shock response. Ann Rev Biochem 55:1151–1191
Lindquist S, Craig EA (1988) The heat shock proteins. Annu Rev Genet 22:631–677
Lovell SC, Davis IW, Arendall WB 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC (2002) Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins 50(3):437–450
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with Folin-phenol reagent. J Biol Chem 193:265–275
Macario AJL, Dugan CB, Conway de Macario E (1991) A dnaK homolog in the archaebacterium Methanosarcina mazei S-6. Gene 108:133–137
Mogk A, Deuerling E, Vorderwulbecke S, Vierling E, Bukau B (2003) Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol 50:585–595
Nimura K, Yoshikawa H, Takahashi H (1994) Identification of dnaK multigene family in Synechococcus sp. strain PCC 7942. Biochem Biophys Res Commun 201:466–471
Ormeno-Orrillo E, Vinuesa P, Zuniga-Davila D, Martinez-Romero E (2006) Molecular diversity of native bradyrhizobia isolated from Lima bean (Phaseolus lunatus L.) in Peru. Syst Appl Microbiol 29:253–262
Perrody E, Cirinesi AM, Desplats C, Keppel F, Schwager F, Tranier S, Georgopoulos C, Genevaux P (2012) A bacteriphage encoded J-domain protein interacts with the DnaK/Hsp70 chaperone and stabilizes the heat shcok factor σ32 of Escherichia coli. PLOS Genetics 8(11):e1003037
Pontius J, Richelle J, Wodak SJ (1996) Deviations from standard atomic volumes as a quality measure for protein crystal structures. J Mol Biol 264(1):121–136
Roland L, Bowie JU, Eisenberg D (1992) Assessment of protein models with three- dimensional profiles. Nature 356:83–85
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schulz A, Schumann W (1996) hrcA, the first gene of the Bacillus subtilis dnaK operon encodes a negative regulator of class I heat shock genes. J Bacteriol 178:1088–1093
Seaton BL, Vickery LE (1994) A gene encoding a new Dnak/hsp70 homolog in Escherichia coli. Proc Natl Acad Sci U S A 91:2066–2070
Sharma RS, Mohmad A, Babu CR (2002) Diversity among rhizobiophages from rhizospheres of legumes inhabiting three ecogeographical regions of India. Soil Biol Biochem 34:965–973
Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V et al (2010) PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res 38:161–166
Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39(Database issue):D561–8
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci 101:11030–11035
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Ward-Rayney N, Rainey FA, Stackebrandt E (1997) The presence of a dnaK (HSP70) multigene family in members of the orders Planctomycetales and Verrucomicrobiales. J Bacteriol 179:6360–6366
Zahran HH (1999) Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol Mol Biol Rev 63:968–989
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jaiswal, S.K., Singh, V.K., Vaishampayan, A. et al. DnaK protein interaction of phage marked Bradyrhizobium of soybean. Ann Microbiol 64, 1535–1542 (2014). https://doi.org/10.1007/s13213-013-0796-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-013-0796-5