Abstract
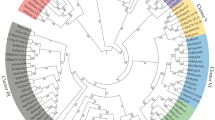
Eukaryotic genomes encode multiple 70-kDa heat-shock proteins (HSP70s). The Saccharomyces cerevisiae HSP70 family is comprised of eight members. Here we present the nucleotide sequence of the SSA3 and SSB2 genes, completing the nucleotide sequence data for the yeast HSP70 family. We have analyzed these yeast sequences as well as 29 HSP70s from 24 additional eukaryotic and prokaryotic species. Comparison of the sequences demonstrates the extreme conservation of HSP70s; proteins from the most distantly related species share at least 45% identity and more than one-sixth of the amino acids are identical in the aligned region (567 amino acids) among all proteins analyzed. Phylogenetic trees constructed by two independent methods indicate that ancient molecular and cellular events have given rise to at least four monophyletic groups of eukaryotic HSP70 proteins. Each group of evolutionarily similar HSP70s shares a common intracellular localization and is presumed to be comprised of functional homologues; these include heat-shock proteins of the cytoplasm, endoplasmic reticulum, mitochondria, and chloroplasts. HSP70s localized in mitochondria and plastids are most similar to the DnaK HSP70 homologues in purple bacteria and cyanobacteria, respectively, which is consistent with the proposed prokaryotic origin of these organelles. The analyses indicate that the major eukaryotic HSP70 groups arose prior to the divergence of the earliest eukaryotes, roughly 2 billion years ago. In some cases, as exemplified by the SSA genes encoding the cytoplasmic HSP70s of S. cerevisiae, more recent duplication events have given rise to subfamilies within the major groups. The S. cerevisiae SSB proteins comprise a unique subfamily not identified in other species to date. This subfamily appears to have resulted from an ancient gene duplication that occurred at approximately the same time as the origin of the major eukaryotic HSP70 groups.
Similar content being viewed by others
References
Atschul SF, Gish W, Miller W, Meyers EW, Lipmann DJ (1990) Basic local alignment tool. J Mol Biol 215:403–410
Boorstein W, Craig EA (1990a) Structure and regulation of the SSA4 HSP70 gene from Saccharomyces cerevisiae. Mol Cell Biol 10:3262–3267
Boorstein W, Craig EA (1990b) Transcriptional regulation of SSA3, an HSP70 gene from Saccharomyces cerevisiae. Mol Cell Biol 10:3262–3267
Bryant D (1992) Puzzles of chloroplast ancestry. Curr Biol 2: 240–242
Cedergren R, Gray MW, Abel Y, Sankoff D (1988) The evolutionary relationships among known life forms. J Mol Evol 28:98–112
Chappell TG, Konforti BB, Schmid SL, Rothman JE (1987) The ATPase core of a clathrin uncoating protein. J Biol Chem 262:746–751
Craig EA, Kang PJ, Boorstein W (1990) A review of the role of 70 kDa heat shock proteins in protein translocation across membranes. Antonie van Leeuwenhoek 58:137–146
Craig EA (1985) The heat shock response. CRC Crit Rev Biochem 18:239–280
Craig EA (1989) Essential roles of 70kDa heat inducible genes. Bioessays 11:48–52
Craig EA, Jacobsen K (1984) Mutations of the heat inducible 70 kilodalton genes of yeast confer temperature sensitive growth. Cell 38:841–849
Craig EA, Jacobsen K (1985) Mutations in cognate gene of Saccharomyces cerevisiae HSP70 result in reduced growth rates at low temperatures. Mol Cell Biol 5:3517–3524
Craig EA, Kramer J, Kosic-Smithers J (1987) SSCI, a member of the 70-kDa heat shock protein multigene family of Saccharomyces cerevisiae, is essential for growth. Proc Natl Acad Sci USA 84:4156–4160
Craig EA, Kramer J, Shilling J, Werner-Washburne M, Holmes S, Kosic-Smither J, Nicolet CM (1989) SSCI, an essential member of the S. cerevisiae HSP70 multigene family, encodes a mitochondrial protein. Mol Cell Biol 9:3000–3008
Craig EA, McCarthy BJ, Wadsworth S (1979) Sequence organization of two recombinant plasmids containing genes for the major heat shock-induced protein of Drosophila melanogaster. Cell 16:575–588
Dayhoff MO, Schwartz RM, Orcutt BC (1978) A model of evolutionary change in proteins. In: Dayhoff MO (ed) Atlas of protein sequence and structure. Silver Spring, The National Biomedical Research Foundation, pp 345–358
Dean N, Pelham H (1990) Recycling of proteins from the Golgi compartment to the ER in yeast. J Cell Biol 111:369–377
Denhardt D (1966) A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun 23: 641–646
Deshaies R, Koch BD, Werner-Washburne M, Craig EA, Shekman R (1988) A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 332:800–805
Devereux J, Haeberli P, Smithies O (1984) A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res 12:387–395
Doolittle RF, Anderson K, Feng D-F (1989) Estimating the prokaryotic-eukaryotic divergence time from protein sequence. In: Fernholm B, Bremer K, Jornvall H (eds) The hierachy of life. Elsevier, New York, pp 73–85
Doolittle RF, Feng D-F (1990) Nearest neighbor procedure for relating progressively aligned amino acid sequences. In: Doolittle RF (ed) Methods in enzymology: the hierarchy of life. Academic Press, San Diego, pp 659–669
Douglas S, Turner S (1991) Molecular evolution for the origin of plastids from cyanobacterium-like ancestor. J Mol Evol 33:267–273
Ellis RJ, van der Vies SM (1991) Molecular chaperones. Annu Rev Biochem 60:321–347
Engebrecht J, Roeder, GS (1989) Yeast mer1 mutants display reduced levels of meiotic recombination. Genetics 121:237–247
Engebrecht J, Roeder, GS (1990) MERI, a yeast gene required for chromosome pairing and genetic recombination, is induced in meiosis. Mol Cell Biol 10:2379–2389
Feng D-F, Doolittle RF (1990) Progressive alignment and phylogenetic tree construction of protein sequences. In: Doolittle RF (ed) Methods in enzymology: molecular evolution: computer analysis of protein and nucleic acid sequences. Academic Press, San Diego pp 375–387
Fitch WM, Margoliash E (1967) Construction of phylogenetic trees. 155:279–284
Flaherty KM, DeLuca-Flaherty C, McKay DB (1990) Three dimensional structure of the ATPase fragment of a 70K heatshock cognate protein. Nature 346:623–628
Flynn G, Pohl J, Flocco M, Rothman J (1991) Peptide-binding specificity of the molecular chaperone BiP. Nature 353:726–730
Flynn GC, Chappell TG, Rothman JE (1989) Peptide binding and release by proteins implicated as catalysts of protein assembly. Science 245:385–390
Fredman, MN (1984) Algorithms for computing evolutionary similarity measures with length independent gap penalties. Bull Math Biol 46:553–566
Ingolia TD, Slater MJ, Craig EA (1982) Saccharomyces cerevisiae contains a complex multigene family related to the major heat shock-inducible gene of Drosophilia. Mol Cell Biol 2: 1388–1398
Kang P-J, Ostermann J, Shilling J, Neupert W, Craig EA, Pfanner N (1990) Requirement for HSP70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature 348:137–143
Klotz LC, Blanken RL (1981) A practical method for calculating the evolutionary trees from sequence data. J Theor Biol 91: 261–272
Ko K, Bornemisza O, Kourtz L, Ko Z, Plaxton W, Cashmore A (1992) Isolation and characterization of a cDNA clone encoding a cognate 70-kDa heat shock protein of the chloroplast envelope. J Biol Chem 2286–2293
Lindquist S (1986) The heat-shock response. Ann Rev Biochem 55:1151–1191
Lindquist S, Craig EA (1988) The heat-shock proteins. Ann Rev Genet 22:631–677
Martindale D, Gu Z, Csank C (1989) Isolation and complete sequence of the yeast isoleucyl-tRNA synthetase gene (ILS1). Curr Genet 15:99–106
McLaughlin CS, Hartwell LH (1969) A mutant of yeast with a defective methionyl-tRNA synthetase. Genetics 61:557–556
Munro S, Pelham HRB (1987) A C-terminal signal prevents secretion of luminal ER proteins. Cell 48:899–907
Nakazawa N, Harashima S, Oshima Y (1991) AAR2, a gene for splicing pre-mRNA of the MATa1 cistron in cell type control of Saccharomyces cerevisiae. Mol Cell Biol 11:5693–5700
Needleman SB, Wunsch CD (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48:443–453
Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA (1992) The translation machinery and seventy kilodalton heat shock protein cooperate in protein synthesis. Cell 71:97–105
Normington K, Kohno K, Kozutsumi Y, Gething MJ, Sambrook J (1989) S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BIP. Cell 57: 1223–1236
Rose MD, Misra LM, Vogel JP (1989) KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 57:1211–1221
Rubin DM, Mehta AD, Zhu JS, Shoham S, Chen X, Wells QR, Palter KB (1993) Genomic structure and sequence analysis of Drosophila melanogaster hsc70 genes. Gene 128:155–163
Sanders S, Whitfield K, Vogel J, Rose M, Schekman (1992) Sec 61p and BiP directly facilitate polypeptide translocation into the ER. Cell 69:353–366.
Sanger F, Nicklen S, Coulson AR (1977) DNA sequence analysis with chain terminating inhibitors. Proc Natl Acad USA 74: 5463–5467
Slater MR, Craig EA (1989a) The SSA1 and SSA2 genes of the yeast Saccharomyces cerevisiae. Nucleic Acids Res 17:805–806
Slater M, Craig E (1989b) SSB1 heat shock cognate gene of the yeast Saccharomyces cerevisiae. Nucleic Acids Res 17:4891–330
Stone DE, Craig EA (1990) Self-regulation of 70-kilodalton heat shock proteins in Saccharomyces cerevisiae Mol Cell Biol 10:1622–1632
Vogel JP, Misra LM, Rose MD (1990) Loss of BiP function blocks translocation of secretory proteins in yeast. J Cell Biol 110:1885–1895
Werner-Washburne M, Stone DE, Craig EA (1987) Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol 7:2568–2577
Woese CR (1987) Bacterial evolution. Microbiol Rev 51:221–271
Author information
Authors and Affiliations
Additional information
Correspondence to: E.A. Craig
Rights and permissions
About this article
Cite this article
Boorstein, W.R., Ziegelhoffer, T. & Craig, E.A. Molecular evolution of the HSP70 multigene family. J Mol Evol 38, 1–17 (1994). https://doi.org/10.1007/BF00175490
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00175490