Abstract
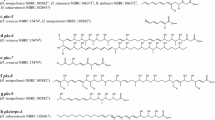
Kitasatospora cheerisanensis KCTC 2395, producing bafilomycin antibiotics belonging to plecomacrolide group, was isolated from a soil sample at Mt. Jiri, Korea. The draft genome sequence contains 8.04 Mb with 73.6% G+C content and 7,810 open reading frames. All the genes for aerial mycelium and spore formations were confirmed in this draft genome. In phylogenetic analysis of MurE proteins (UDP-N-acetylmuramyl-L-alanyl-D-glutamate:DAP ligase) in a conserved dcw (division of cell wall) locus, MurE proteins of Kitasatospora species were placed in a separate clade between MurEs of Streptomyces species incorporating LL-diaminopimelic acid (DAP) and MurEs of Saccharopolyspora erythraea as well as Mycobacterium tuberculosis ligating meso-DAP. From this finding, it was assumed that Kitasatospora MurEs exhibit the substrate specificity for both LL-DAP and meso-DAP. The bafilomycin biosynthetic gene cluster was located in the left subtelomeric region. In 71.3 kb-long gene cluster, 17 genes probably involved in the biosynthesis of bafilomycin derivatives were deduced, including 5 polyketide synthase (PKS) genes comprised of 12 PKS modules.
Similar content being viewed by others
References
Barbe, V., Bouzon, M., Mangenot, S., Badet, B., Poulain, J., Segurens, B., Vallenet, D., Marlière, P., and Weissenbach, J. 2011. Complete genome sequence of Streptomyces cattleya NRRL 8057, a producer of antibiotics and fluorometabolites. J. Bacteriol. 193, 5055–5056.
Basavannacharya, C., Robertson, G., Munshi, T., Keep, N.H., and Bhakta, S. 2010. ATP-dependent MurE ligase in Mycobacterium tuberculosis: biochemical and structural characterisation. Tuberculosis 90, 16–24.
Bentley, S.D., Chater, K.F., Cerdeño-Tárraga, A.M., Challis, G.L., Thomson, N.R., James, K.D., Harris, D.E., Quail, M.A., Kieser, H., Harper, D., et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417,141–147.
Bignell, D.R., Seipke, R.F., Huguet-Tapia, J.C., Chambers, A.H., Parry, R.J., and Loria, R. 2010. Streptomyces scabies 87-22 contains a coronafacic acid-like biosynthetic cluster that contributes to plant-microbe interactions. Mol. Plant Microbe Interact. 23, 161–175.
Chater, K.F. and Chandra, G. 2006. The evolution of development in Streptomyces analysed by genome comparisons. FEMS Microbiol. Rev. 30, 651–672.
Chung, Y.R., Sung, K.C., Mo, H.K., Son, D.Y., Nam, J.S., Chun, J., and Bae, K.S. 1999. Kitasatospora cheerisanensis sp. nov., a new species of the genus Kitasatospora that produces an antifungal agent. Int. J. Syst. Bacteriol. 49, 753–758.
Claessen, D., de Jong, W., Dijkhuizen, L., and Wösten, H.A.B. 2006. Regulation of Streptomyces development: reach for the sky! Trends Microbiol. 14, 313–319.
Delcher, A.L., Salzberg, S.L., and Phillippy, A.M. 2003. Using MUMmer to identify similar regions in large sequence sets. Curr. Protoc. Bioinformatics Chap.10. doi: 10.1002/0471250953.bi1003s00.
den Hengst, C.D., Tran, N.T., Bibb, M.J., Chandra, G., Leskiw, B.K., and Buttner, M.J. 2010. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 78, 361–379.
Disz T., Akhter S., Cuevas D., Olson R., Overbeek R., Vonstein V., Stevens R., and Edwards, R.A. 2010. Accessing the SEED genome databases via Web services API: tools for programmers. BMC Bioinformatics 11, 319.
Hwang, J.Y., Kim, H.S., Kim, S.H., Oh, H.R., and Nam, D.H. 2013. Organization and characterization of a biosynthetic gene cluster for bafilomycin from Streptomyces griseus DSM 2608. AMB Express 3, 24.
Hwang, J.Y., Kim, S.H., Oh, H.R., Cho, Y.J., Chun, J., Chung, Y.R., and Nam, D.H. 2014. Draft genome sequence of Kitasatospora cheerisanensis KCTC 2395 producing plecomacrolide against phytopathogenic fungi. Genome Announc. 2, e00604–4.
Ichikawa, N., Oguchi, A., Ikeda, H., Ishikawa, J., Kitani, S., Watanabe, Y., Nakamura, S., Katano, Y., Kishi, E., Sasagawa, M., et al. 2010. Genome sequence of Kitasatospora setae NBRC 14216: An evolutionary snapshot of the family Streptomycetaceae. DNA Res. 17, 393–406.
McGroty, S.E., Pattaniyil, D.T., Patin, D., Blanot, D., Ravichandran, A.C., Suzuki, H., Dobson, R.C., Savka, M.A., and Hudson, A.O. 2013. Biochemical characterization of UDP-N-acetylmuramoyl-L-alanyl-D-glutamate: meso-2,6-diaminopimelate ligase (MurE) from Verrucomicrobium spinosum DSM 4136(T.). PLoS One 8, e66458.
Moon, S.S., Hwang, W.H., Chung, Y.R., and Shin, J. 2003. New cytotoxic bafilomycin C1-amide produced by Kitasatospora cheerisanensis. J. Antibiot. 56, 856–861.
Ohnishi, Y., Ishikawa, J., Hara, H., Suzuki, H., Ikenoya, M., Ikeda, H., Yamashita, A., Hattori, M., and Horinouchi, S. 2008. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190, 4050–4060.
Oliynyk, M., Samborskyy, M., Lester, J.B., Mironenko, T., Scott, N., Dickens, S., Haydock, S.F., and Leadlay, P.F. 2007. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat. Biotechnol. 25, 447–453.
Omura, S., Ikeda, H., Ishikawa, J., Hanamoto, A., Takahashi, C., Shinose, M., Takahashi, Y., Horikawa, H., Nakazawa, H., Osonoe, T., et al. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 98, 12215–12220.
Omura, S., Iwai, Y., Takahashi, Y., Kojima, K., Otoguro, K., and Oiwa, R. 1981. Type of diaminopimelic acid different in aerial and vegetative mycelia of setamycin-producing actinomycete KM-6054. J. Antibiot. 34, 1633–1634.
Omura, S., Takahashi, Y., Iwai, Y., and Tanaka, H. 1982. Kitasatospora, a new genus of the order Actinomycetales. J. Antibiot. 35, 1013–1019.
Shanmugam, A. and Natarajan, J. 2012. Comparative modeling of UDP-N-acetylmuramoyl-glycyl-D-glutamate-2, 6-diaminopimelate ligase from Mycobacterium leprae and analysis of its binding features through molecular docking studies. J. Mol. Model. 18, 115–125.
Smith, C.A. 2006. Structure, function and dynamics in the mur family of cell wall ligases. J. Mol. Biol. 362, 640–655.
Tae, H., Sohng, J.K., and Park, K. 2009. Development of an analysis program of type I polyketide synthase gene clusters using homology search and profile hidden Markov model. J. Microbiol. Biotechnol. 19, 140–146.
Takahashi, Y., Iwai, Y., and Omura, S. 1983. Relationship between cell morphology and the types of diaminopimelic acid in Kitasatospora setalba. J. Gen. Appl. Microbiol. 29, 459–465.
Tatusov, R.L., Galperin, M.Y., Natale, D.A., and Koonin, E.V. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36.
Tatusov, R.L., Koonin, E.V., and Lipman, D.J. 1997. A genomic perspective on protein families. Science 278, 631–637.
Zhang, W., Fortman, J.L., Carlson, J.C., Yan, J., Liu, Y., Bai, F., Guan, W., Jia, J., Matainaho, T., Sherman, D.H., et al. 2013. Characterization of the bafilomycin biosynthetic gene cluster from Streptomyces lohii. Chembiochem 14, 301–306.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hwang, J.Y., Kim, S.H., Oh, H.R. et al. Analysis of a draft genome sequence of Kitasatospora cheerisanensis KCTC 2395 producing bafilomycin antibiotics. J Microbiol. 53, 84–89 (2015). https://doi.org/10.1007/s12275-015-4340-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-015-4340-0