Abstract
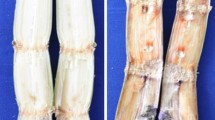
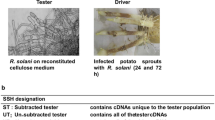
Bacterial wilt, one of the major diseases in ginger is caused by Ralstonia solanacearum and lack of resistant genotype adds constraints to the crop improvement programs and for the management of bacterial wilt. Curcuma amada is a potential donor for bacterial wilt resistance to Zingiber officinale, if the exact mechanism of resistance is understood. In this study, a PCR-based suppression subtractive hybridization (SSH) was used to identify C. amada genes that are differentially and early expressed in response to the R. solanacearum infection compared to Z. officinale. A forward subtracted library of differentially expressed genes was synthesized by cloning DNA fragments (300–1,200 bp) from subtracted sample and sequenced. Upon computational analysis of the 150 SSH clones sequenced, 119 produced suitable sequences of high homology with the genes of known functions. These were classified into three potential functional categories: proteins involved in biological processes (31 %), cellular component (43 %) and molecular function (26 %). Among these differentially expressed genes, sequences coding for putative proteins related to defense response (high homology between 85 and 100 % identity) were, cell wall-associated hydrolase, xyloglucan transglycosylase (XTG), cytochrome P 450, metallocarboxypeptidase inhibitor, peroxiredoxin, nitrilase-associated protein, leucine-rich protein (LRR) and glutathione-S-transferase (GST). The three transcripts were discriminative: expression of LRR, GST and XTG was much higher in resistant species (C. amada) than in susceptible species (Z. officinale) at every time point. The isolated ESTs reported here contribute to improve our understanding about this plant–pathogen interaction.



Similar content being viewed by others
References
Aslam M, Sinha VB, Singh RK, Anandhan S, Pande V, Ahmed Z (2010) Isolation of cold stress-responsive genes from Lepidium latifolium by suppressive subtraction hybridization. Acta Physiol Plant 32:205–210
Belkhadir Y, Subramaniam R, Dangl JL (2004) Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr Opin Plant Biol 7(4):391–399
Cantu D, Vicente AR, Labavitch JM, Bennett AB, Powell ALT (2008) Strangers in the matrix: plant cell walls and pathogen susceptibility. Tren Plant Sci 13:610–617
Dangle JL, Jones JDG (2001) Plant pathogens and integrated defence response to infection. Nature 411:826–833
de Souza CRB, Brı′gida ABS, Santos RCD, de Nazare′ Monteiro Costa C, Darnet SH, Harada ML (2011) Identification of sequences expressed during compatible black pepper—Fusarium solani f. sp. piperis interaction. Acta Physiol Plant 33:2553–2560
Dean JD, Goodwin PH, Hsiang T (2005) Induction of glutathione-S-transferase genes of Nicotiana benthemiana following infection by Colletotrichum destructivum and C. orbiculare and involvement of one in resistance. J Exp Bot 56:1525–1533
Diatchenko L, Lau YFC, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA 93:6025–6030
Fan F, Li X-W, Wu Y-M, Xia Z-S, Li J–J, Zhu W, Liu J-X (2010) Differential expression of expressed sequence tags in alfalfa roots under aluminum stress. Acta Physiol Plant 33(2):539–546
Fekete C, Fung RWM, Szabo′d Z, Qiu W, Chang L, Schachtman DP, Kova′cs LG (2009) Up-regulated transcripts in a compatible powdery mildew–grapevine interaction. Plant Physiol Biochem 47:732–738
Ghose K, Dey S, Barton H, Loake GJ, Basu D (2008) Differential profiling of selected defence-related genes induced on challenge with Alternaria brassiciola in resistant white mustard and their comparative expression pattern in susceptible India mustard. Mol Plant Pathol 9:763–775
Glazebrook J (2001) Genes controlling expression of defense responses in Arabidopsis-2001 status. Curr Opin Plant Biol 4:301–308
Gor MC, Ismail I, Mustapha WAW, Zainal Z, Noor NM, Othman R, Hussein ZAM (2010) Identification of cDNAs for jasmonic acid responsive genes in polygonum minus roots by suppression subtractive hybridization. Acta Physiol Plant 33:283–294
Hammond-Kosack KE, Jones JDG (1996) Resistance gene dependent plant defense responses. Plant Cell 8:1773–1791
Hirao T, Fukatsu E, Watanabe A (2012) Characterization of resistance to pine wood nematode infection in Pinus thunbergii using suppression subtractive hybridization. BMC Plant Biol 12:13. doi:10.1186/1471-2229-12-13
Kavitha PG, Thomas G (2008) Population genetic structure of the clonal plant Zingiber zerumbet (L.) Smith (Zingiberaceae), a wild relative of cultivated ginger, and its response to Pythium aphanidermatum. Euphytica 160:89–100
Kelman A (1954) The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on a tetrazolium medium. Phytopathol 44:693–695
Kim YC, Kim SY, Paek KH, Choi D, Park JM (2006) Suppression of CaCYP1, a novel cytochrome P450 gene, compromises the basal pathogen defense response of pepper plants. Biochem Biophys Res Commun 345:638–645
Kumar A, Hayward AC (2005) Bacterial diseases of ginger and their control. In: Ravindran PN, Babu KN (eds) Monograph on Ginger. CRC Press, USA, pp 341–366
Kumar A, Bhai RS, Sasikumar B, Anandaraj M, Parthasarathy VA (2006) Curcuma amada Roxb. A bacterial wilt evading species in Zingiberaceae—A potential source of valuable genes for bacterial wilt resistance. In: The 4th International Bacterial wilt symposium, 17–20th July 2006. The Lakeside conference centre, Central Science Laboratory, York, UK
Latha AM, Rao KV, Reddy VD (2005) Production of transgenic plants resistant to leaf blast disease in finger millet (Eleusine coracana (L.) Gaertn.). Plant Sci 169:657–667
Luo M, Kong X, Huo N, Zhou R, Jia J (1990) Gene expression profiling related to powdery mildew resistance in wheat with the method of suppression subtractive hybridization. Chin Sci Bull 47:1990–1994
Mukherjee AK, Lev S, Gepstein S, Horwitz BA (2009) A compatible interaction of Alternaria brassicicola with Arabidopsis thaliana ecotype DiG: evidence for a specific transcriptional signature. BMC Plant Biol 9:31
Nascimento SB, Cascardo JCM, de Menezes IC, Duarte MLR, Darnet SH, Harada ML, de Souza CRB (2009) Identifying sequences potentially related to resistance response of Piper tuberculatum to Fusarium solani f. sp. piperis by suppression subtractive hybridization. Prot Pept Lett 12:1429–1434
Nishitani K (1998) Construction and restructuring of the cellulose-xyloglucan framework in the apoplast as mediated by the xyloglucan related protein family: a hypothetical scheme. J Plant Res 111:159–166
Noctor G, Foyer CH (1998) Ascorbate and glutathione: keeping active oxygen under control. Ann Rev Plant Physiol Plant Mol Biol 49:249–279
Oh BJ, Ko MK, Kim YS, Kim KS, Kostenyuk I, Kee HK (1999) A cytochrome P450 gene is differentially expressed in compatible and incompatible interactions between pepper (Capsicum annuum) and the anthracnose fungus, Colletotrichum gloeosporioides. Mol Plant Microbe Interact 12:1044–1052
Prasath D, El-Sharkawy I, Tiwary KS, Jayasankar S, Sherif S (2011) Cloning and characterization of PR5 gene from Curcuma amada and Zingiber officinale in response to Ralstonia solanacearum infection. Plant Cell Rep 30(10):1799–1809
Ros B, Thummler F, Wenzel G (2004) Analysis of differentially expressed genes in a susceptible and moderately resistant potato cultivar upon Phytophthora infestans infection. Mol Plant Pathol 5:191–201
Schlink K (2010) Down-regulation of defense genes and resource allocation into infected roots as factors for compatibility between Fagus sylvatica and Phytophthora citricola. Func Integ Genom 10:253–264
Sharmin S, Azam MS, Islam MS, Sajib AA, Mahmood N, Hasan AM, Ahmed R, Sultana K, Khan H (2012) Xyloglucan endotransglycosylase/hydrolase genes from a susceptible and resistant jute species show opposite expression pattern following Macrophomina phaseolina infection. Comm Integ Biol 5(6):1–9
Stacy RAP, Munthe E, Steinum T, Sharma B, Aalen RB (1996) A peroxiredoxin antioxidant is encoded by a dormancy-related gene, Per1, expressed during late development in the aleurone and embryo of barley grains. Plant Mol Biol 31:1205–1216
Thompson GA, Goggin FL (2006) Transcriptomics and functional genomics of plant defense induction by phloem-feeding insects. J Exp Bot 57:755–766
Tian ZD, Liu J, Xie CH (2003) Isolation of resistance related-genes to Phytophthora infestans with suppression subtractive hybridization in the R-gene free potato. Acta Genet Sin 30:597–605
Urbanowicz BR, Catala C, Irwin D, Wilson DB, Ripoll DR, Rose JK (2007) A tomato endo-beta-1,4-glucanase, SlCel9C1, represents a distinct subclass with a new family of carbohydrate binding modules (CBM49). J Biol Chem 282:12066–12074
Veronese P, Ruiz MT, Coca MA, Hernandez-Lopez A, Lee H, Ibeas JI (2003) In defense against pathogens. Both plant sentinels and foot soldiers need to know the enemy. Plant Physiol 131:1580–1590
Xu P, Narasimhan ML, Samson T, Coca MA, Huh G, Zhou J, Martin GB, Hasegawa PM, Bressan RA (1998) A nitrilase-like protein interacts with GCC box DNA-binding proteins involved in ethylene and defense responses. Plant Physiol 118(3):867–874
Yang T, Shah J, Klessig DF (1997) Signal perception and transduction in plant defense responses. Gen Dev 11:1621–1639
Yang GP, Ross DT, Kang WW, Brown PO, Weigel RJ (1999) Combining SSH and cDNA microarray for rapid identification of differentially expressed genes. Nucl Acids Res 27:1517–1523
Zuo KJ, Qin J, Zhao JY, Ling H, Zhang LD, Cao YF, Tang KX (2007) Over-expression of GbERF2 transcription factors in tobacco enhanced brown spots disease resistance by activating expression of downstream genes. Gene 391:80–90
Acknowledgments
Funding from Indian Council of Agricultural Research (ICAR) as ORP (PhytoFuRa) is gratefully acknowledged. Authors are grateful to Director, IISR; Head, Crop Improvement and Biotechnology, IISR; DISC, IISR; Head, Crop Protection, SBI, Coimbatore for technical help and support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Kuzniak-Gebarowska.
Rights and permissions
About this article
Cite this article
Prasath, D., Suraby, E.J., Karthika, R. et al. Analysis of differentially expressed genes in Curcuma amada and Zingiber officinale upon infection with Ralstonia solanacearum by suppression subtractive hybridization. Acta Physiol Plant 35, 3293–3301 (2013). https://doi.org/10.1007/s11738-013-1362-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11738-013-1362-2