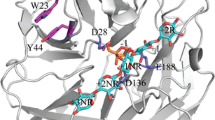
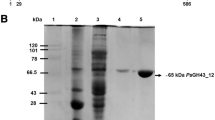
Abstract
Arabinofuranosidase Abf43A from Bacillus sp. BP-7 is a newly discovered arabinoxylan arabinofuranohydrolase (AXH). It is a modular enzyme comprised of a GH43 catalytic domain and a carbohydrate-binding module of family CBM6. Recombinant Abf43A showed high activity on arabinoxylans, being rye arabinoxylan the preferred substrate on which the purified enzyme exhibited a K m of 10.6 ± 3.3 mg/ml and a V max of 29.2 ± 3.4 U/mg. Thin-layer chromatography analysis of hydrolysis products showed arabinose as the only sugar released by the enzyme from its substrates. The GH43 and CBM6 modules of the enzyme were individually cloned and expressed in Escherichia coli. While the isolated catalytic GH43 module did not show hydrolytic activity, the purified CBM6 bound to soluble arabinoxylan in affinity gel electrophoresis analysis. Evaluation of cooperative activity of arabinofuranosidase Abf43A with xylanases from families GH10, GH11, and GH30, (Xyn10A, Xyn11E, and Xyn30D from Paenibacillus barcinonensis) on arabinoxylan depolymerization revealed that the studied enzyme showed synergism with Xyn11E, a 2.54-fold increase in the amount of sugars released. On the contrary, Abf43A did not show synergism with the xylanases of families GH10 or GH30 evaluated. The enzyme characterized contributes to understanding the role of this class of enzymes in the catalytic depolymerization of arabinoxylans and their potential for the production of valuable xylooligosaccharides from these abundant plant polymers.





Similar content being viewed by others
References
Biely P, Vršanská M, Tenkanen M, Kluepfel D (1997) Endo-beta-1,4-xylanase families: differences in catalytic properties. J Biotechnol 57:151–166. doi:10.1016/S0168-1656(97)00096-5
Boraston AB, Bolam DN, Gilbert HJ, Davies GJ (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382:769–781. doi:10.1042/BJ20040892
Bourgois TM, Van Craeyveld V, Van Campenhout S, Courtin CM, Delcour JA, Robben J, Volckaert G (2007) Recombinant expression and characterization of XynD from Bacillus subtilis subsp. subtilis ATCC 6051: a GH 43 arabinoxylan arabinofuranohydrolase. Appl Microbiol Biotechnol 75:1309–1317. doi:10.1007/s00253-007-0956-2
Broekaert WF, Courtin CM, Verbeke K, Van de Wiele T, Verstraete W, Delcour JA (2011) Prebiotic and other health-related effects of cereal-derived arabinoxylans, arabinoxylan-oligosaccharides, and xylooligosaccharides. Crit Rev Food Sci Nutr 51:178–194. doi:10.1080/10408390903044768
Burstein T, Shulman M, Jindou S, Petkun S, Frolow F, Shoham Y, Bayer EA, Lamed R (2009) Physical association of the catalytic and helper modules of a family-9 glycoside hydrolase is essential for activity. FEBS Lett 583:879–884. doi:10.1016/j.febslet.2009.02.013
Chiriac AI, Cadena EM, Vidal T, Torres AL, Diaz P, Pastor FIJ (2010) Engineering a family 9 processive endoglucanase from Paenibacillus barcinonensis displaying a novel architecture. Appl Microbiol Biotechnol 86:1125–1134. doi:10.1007/s00253-009-2350-8
Gallardo O, Fernández-Fernández M, Valls C, Valenzuela SV, Roncero MB, Vidal T, Díaz P, Pastor FIJ (2010) Characterization of a family GH5 xylanase with activity on neutral oligosaccharides and evaluation as a pulp bleaching aid. Appl Environ Microbiol 76:6290–6294. doi:10.1128/AEM.00871-10
Goldbeck R, Damásio ARL, Gonçalves TA, Machado CB, Paixão DAA, Wolf LD, Mandelli F, Rocha GJM, Ruller R, Squina FM (2014) Development of hemicellulolytic enzyme mixtures for plant biomass deconstruction on target biotechnological applications. Appl Microbiol Biotechnol 98:8513–8525. doi:10.1007/s00253-014-5946-6
Gonçalves TA, Damásio ARL, Segato F, Alvarez TM, Bragatto J, Brenelli LB, Citadini APS, Murakami MT, Ruller R, Paes Leme AF, Prade RA, Squina FM (2012) Functional characterization and synergic action of fungal xylanase and arabinofuranosidase for production of xylooligosaccharides. Bioresour Technol 119:293–299. doi:10.1016/j.biortech.2012.05.062
Hövel K, Shallom D, Niefind K, Belakhov V, Shoham G, Baasov T, Shoham Y, Schomburg D (2003) Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase. EMBO J 22:4922–4932. doi:10.1093/emboj/cdg494
Juturu V, Wu JC (2012) Microbial xylanases: engineering, production and industrial applications. Biotechnol Adv 30:1219–1227. doi:10.1016/j.biotechadv.2011.11.006
Keasling JD (2010) Manufacturing molecules through metabolic engineering. Science 330:1355–1358. doi:10.1126/science.1193990
Kormelink FJM, Searle-Van Leeuwen MJF, Wood TM, Voragen AGJ (1991) (1,4)-B-d-arabinoxylan arabinofuranohydrolase: a novel enzyme in the bioconversion of arabinoxylan. Appl Microbiol Biotechnol. doi: 10.1007/BF00184692
Lagaert S, Pollet A, Delcour JA, Lavigne R, Courtin CM, Volckaert G (2010) Substrate specificity of three recombinant α-L-arabinofuranosidases from Bifidobacterium adolescentis and their divergent action on arabinoxylan and arabinoxylan oligosaccharides. Biochem Biophys Res Commun 402:644–650. doi:10.1016/j.bbrc.2010.10.075
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi:10.1093/nar/gkt1178
López C, Blanco A, Pastor FIJ (1998) Xylanase production by a new alkali-tolerant isolate of Bacillus. Biotechnol Lett 20:243–246. doi:10.1023/A:1005321701384
Maehara T, Fujimoto Z, Ichinose H, Michikawa M, Harazono K, Kaneko S (2014) Crystal structure and characterization of the glycoside hydrolase family 62 α-L-arabinofuranosidase from Streptomyces coelicolor. J Biol Chem 289:7962–7972. doi:10.1074/jbc.M113.540542
Mangala SL, Kittur FS, Nishimoto M, Sakka K, Ohmiya K, Kitaoka M, Hayashi K (2003) Fusion of family VI cellulose binding domains to Bacillus halodurans xylanase increases its catalytic activity and substrate-binding capacity to insoluble xylan. J Mol Catal B Enzym 21:221–230. doi:10.1016/S1381-1177(02)00226-6
Numan MT, Bhosle NB (2006) Alpha-L-arabinofuranosidases: the potential applications in biotechnology. J Ind Microbiol Biotechnol 33:247–260. doi:10.1007/s10295-005-0072-1
Pitkänen L, Virkki L, Tenkanen M, Tuomainen P (2009) Comprehensive multidetector HPSEC study on solution properties of cereal arabinoxylans in aqueous and DMSO solutions. Biomacromolecules 10:1962–1969. doi:10.1021/bm9003767
Pollet A, Delcour JA, Courtin CM (2010) Structural determinants of the substrate specificities of xylanases from different glycoside hydrolase families. Crit Rev Biotechnol 30:176–191. doi:10.3109/07388551003645599
Prade RA (1996) Xylanases: from biology to biotechnology. Biotechnol Genet Eng Rev 13:101–132
Puls J, Schuseil J (1993) Chemistry of hemicelluloses: relationship between hemicellulose structure and enzymes required for hydrolysis. In: Coughlan MP, Hazlewood. GP (eds) Hemicellulose and Hemicellulases. Portland Press Ltd., pp 1–27
Rasmussen LE, Xu C, Sørensen JF, Nielsen MK, Meyer AS (2012) Enzyme kinetics and identification of the rate-limiting step of enzymatic arabinoxylan degradation. Biochem Eng J 69:8–16. doi:10.1016/j.bej.2012.08.004
Saha BC (2000) Alpha-L-arabinofuranosidases: biochemistry, molecular biology and application in biotechnology. Biotechnol Adv 18:403–423. doi:10.1016/S0734-9750(00)00044-6
Sainz-Polo MA, Valenzuela SV, González B, Pastor FIJ, Sanz-Aparicio J (2014) Structural analysis of glucuronoxylan-specific Xyn30D and its attached CBM35 domain gives insights into the role of modularity in specificity. J Biol Chem 289:31088–31101. doi:10.1074/jbc.M114.597732
Sakka M, Tachino S, Katsuzaki H, van Dyk JS, Pletschke BI, Kimura T, Sakka K (2012) Characterization of Xyn30A and Axh43A of Bacillus licheniformis SVD1 identified by its genomic analysis. Enzym Microb Technol 51:193–199. doi:10.1016/j.enzmictec.2012.06.003
Scheller HV, Ulvskov P (2010) Hemicelluloses. Annu Rev Plant Biol 61:263–289. doi:10.1146/annurev-arplant-042809-112315
Sørensen HR, Pedersen S, Meyer AS (2007) Synergistic enzyme mechanisms and effects of sequential enzyme additions on degradation of water insoluble wheat arabinoxylan. Enzym Microb Technol 40:908–918. doi:10.1016/j.enzmictec.2006.07.026
St John FJ, Dietrich D, Crooks C, Pozharski E, González JM, Bales E, Smith K, Hurlbert JC (2014) A novel member of glycoside hydrolase family 30 subfamily 8 with altered substrate specificity. Acta Crystallogr D Biol Crystallogr 70:2950–2958. doi:10.1107/S1399004714019531
St John FJ, Hurlbert JC, Rice JD, Preston JF, Pozharski E (2011) Ligand bound structures of a glycosyl hydrolase family 30 glucuronoxylan xylanohydrolase. J Mol Biol 407:92–109. doi:10.1016/j.jmb.2011.01.010
Tuck CO, Pérez E, Horváth IT, Sheldon RA, Poliakoff M (2012) Valorization of biomass: deriving more value from waste. Science 337:695–699. doi:10.1126/science.1218930
Urbániková L, Vršanská M, Mørkeberg Krogh KBR, Hoff T, Biely P (2011) Structural basis for substrate recognition by Erwinia chrysanthemi GH30 glucuronoxylanase. FEBS J 278:2105–2116. doi:10.1111/j.1742-4658.2011.08127.x
Valenzuela SV, Diaz P, Pastor FIJ (2014a) Xyn11E from Paenibacillus barcinonensis BP-23: a LppX-chaperone-dependent xylanase with potential for upgrading paper pulps. Appl Microbiol Biotechnol 98:5949–5957. doi:10.1007/s00253-014-5565-2
Valenzuela SV, Diaz P, Pastor FIJ (2010) Recombinant expression of an alkali stable GH10 xylanase from Paenibacillus barcinonensis. J Agric Food Chem 58:4814–4818. doi:10.1021/jf9045792
Valenzuela SV, Diaz P, Pastor FIJ (2012) Modular glucuronoxylan-specific xylanase with a family CBM35 carbohydrate-binding module. Appl Environ Microbiol 78:3923–3931. doi:10.1128/AEM.07932-11
Valenzuela SV, Valls C, Roncero MB, Vidal T, Diaz P, Pastor FIJ (2014b) Effectiveness of novel xylanases belonging to different GH families on lignin and hexenuronic acids removal from specialty sisal fibres. J Chem Technol Biotechnol 89:401–406. doi:10.1002/jctb.4132
Valls C, Vidal T, Gallardo O, Diaz P, Pastor FIJ, Roncero MB (2010) Obtaining low-HexA-content cellulose from eucalypt fibres: which glycosil hydrolase family is more efficient? Carbohydr Polym 80:154–160. doi:10.1016/j.carbpol.2009.11.006
Van Laere KM, Beldman G, Voragen AG (1997) A new arabinofuranohydrolase from Bifidobacterium adolescentis able to remove arabinosyl residues from double-substituted xylose units in arabinoxylan. Appl Microbiol Biotechnol 47:231–235. doi:10.1007/s002530050918
Vandermarliere E, Bourgois TM, Winn MD, van Campenhout S, Volckaert G, Delcour JA, Strelkov SV, Rabijns A, Courtin CM (2009) Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family. Biochem J 418:39–47. doi:10.1042/BJ20081256
Vardakou M, Katapodis P, Topakas E, Kekos D, Macris BJ, Christakopoulos P (2004) Synergy between enzymes involved in the degradation of insoluble wheat flour arabinoxylan. Innov Food Sci Emerg Technol 5:107–112. doi:10.1016/S1466-8564(03)00044-4
Acknowledgments
This work was partially supported by the Spanish Ministry of Economy and Competitiveness, project ref. CTQ2013-48995-C2-2-R; and Xarxa de Referència en Biotecnologia (XRB).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Valls, A., Diaz, P., Pastor, F.I.J. et al. A newly discovered arabinoxylan-specific arabinofuranohydrolase. Synergistic action with xylanases from different glycosyl hydrolase families. Appl Microbiol Biotechnol 100, 1743–1751 (2016). https://doi.org/10.1007/s00253-015-7061-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-7061-8