Abstract
The 13th International Wheat Genetics Symposium (IWGS) is being held in the year I begin my phased retirement, marking a career of 40 years in wheat genetics, beginning with a postdoctoral fellowship in 1973 with Ernie Sears and Gordon Kimber at Columbia, Missouri, then the premier center for wheat chromosome research. I was fortunate to have won a DF Jones fellowship for my research proposal, “Exploration and application of the Quinacrine and Giemsa staining technique in the genus Triticum” that led to the cytogenetic identification of wheat and rye chromosomes (Gill and Kimber 1974a, b). In 1973, I also attended, for the first time, the meetings of the 4th IWGS in Columbia, Missouri, and was in awe of the research presentations and heated discussions on wheat evolution. In 1979, I established my own research group and laboratory at Kansas State University focusing on wheat chromosome mapping and manipulation for crop improvement under the auspices of Wheat Genetics Resource Center (reviewed in Raupp and Friebe, Plant Breed Rev 37:1–34, 2013). Among the first visitors to my laboratory were Takashi Endo, then at Nara University, Japan, and Chen Peidu, from Nanjing Agricultural University, and presented this research at the 6th IWGS in Kyoto, Japan. Therefore, it is a special feeling to be returning to Japan for a farewell presentation. My intent is to briefly review the history of wheat chromosome research and how our laboratory played a role in advancing wheat chromosome analysis leading to the chromosome survey sequencing paper utilizing telosomic stocks (IWGSC, Science 345:285–287, 2014).
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
Laying the Foundation of Wheat Chromosome Research: Genome Analyzer Method
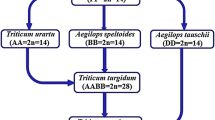
By 1915, botanists had described three classes of cultivated wheats, the one-seeded monococcum (Triticum monococcum L.), the two-seeded emmer (T. turgidum L.), and dinkel (T. aestivum L.). The one-seeded wild relative of monococcum was reported in Greece and Anatolia between 1834 and 1884. Aaronsohn discovered the two-seeded wild relative of emmer in 1910 in Lebanon, Syria, Jordan, and Israel. Therefore, it was well accepted, as Candolle had suggested in 1886, that since wild wheats grow in the Euphrates basin then wheat cultivation must have originated there. Between 1918 and 1924, Sakamura (1918) and his colleague Hitoshi Kihara (1919), at Hokkaido Imperial University in Japan, and Karl Sax (1922), at Harvard University, reported their classic studies on the genetic architecture of the three groups of wheats (Fig. 7.1). Sax (1922) and Kihara (1924) analyzed meiosis in wheat species and hybrids and were the first to establish the basic chromosome number of seven and document polyploidy in the wheat group. This method of delineating species evolutionary relationships based on chromosome pairing affinities in interspecific hybrids came to be called as the genome analyzer method (Kihara 1954; see also Fig. 7.1 in Gill et al. 2006). These were exciting observations and established polyploidy as a major macrospeciation process and wheat as a great polyploidy genetic model. These interploidy wheat hybrids of course could also be exploited in plant breeding for interspecific gene transfers (McFadden 1930; Gill and Raupp 1987).
Meiotic metaphase I chiasmate pairing in wheat parents and F1 hybrids. The F1 hybrid between einkorn and dicoccum showed 7″ and 7′, indicating that they share one set of chromosomes in common, and it was called a genome. The second genome in dicoccum was called the B genome. The F1 hybrids between dicoccum and dinkel showed 14″ and 7′ indicating that they share the AB genomes in common, and dinkel wheat had a third genome that was later identified as D genome
The crowning achievement of the genome analyzer method was the identification of the D-genome donor of wheat (Kihara 1944; McFadden and Sears 1946) and the production of synthetic wheat (McFadden and Sears 1944). These discoveries are fueling a second green revolution (reviewed in Gill et al. 2006). Kimber practiced the genome analyzer method with passion and developed some quantitative models for measuring genomic affinities. Application of Giemsa staining methods to meiotic preparations allowed the measurement of pairing potential of specific chromosomes, however, perfectly homologous chromosomes may suffer structural aberrations and lose the ability to pair (Gill and Chen 1987; Naranjo et al. 1987). Obviously, the genome analyzer method had reached its limitations, but meiotic pairing analysis remains an important method for monitoring chiasmate pairing and the potential of genetic transfers in interspecific hybrids.
Laying the Foundation for Cytogenetic and Genome Mapping: The Wheat Aneuploid Stocks
Sears (1939, 1954) isolated a range of aneuploid stocks in Chinese Spring wheat that were widely used by the wheat genetics community to dissect and map the polyploid genome of wheat (Fig. 7.2). The monosomic and telosomics were useful for chromosome and arm mapping of genes, and for producing intervarietal substitution lines for the mapping of quantitative traits (Law 1966). Wheat nulli-tetrasomics were useful for comparative genetic mapping and determining homoeologous relationships among chromosomes of different genomic origin (Sears 1966). O’Mara (1940) developed an aneuploid approach for mapping alien chromosomes added to wheat.
Molecular cytogenetic mapping of wheat phenotypes and the genome. Steps in targeted physical mapping of genes in wheat. Absence of the marker in the critical nullisomic-tetrasomic, long arm ditelosomic deletion stocks, delS1 and delS2, and presence of the marker in the short arm ditelosomic, deletion stocks delS3 and delS4 maps the gene to the chromosome bin 3, which is defined by the deletion interval spanned by delS3 and delS4 (chromosome segments missing in the aneuploid stocks are shown in hatching) (Originally published in Faris et al. (2002))
Gametocidal chromosomes were used to isolate deletion stocks (Endo and Gill 1996) for mapping traits and ESTs (expressed sequence tags) into deletion bins (Qi et al. 2004). This extensive mapping of chromosomes revealed structural and functional differentiation along the centromere and telomere axis and the unequal distribution of genes and recombination towards the distal ends of chromosomes.
The individual telosomic chromosomes, reported by Sears and Sears (1978), have been sequenced (IWGSC 2014) using chromosome genomic methods pioneered by Dolezel and his group (reviewed elsewhere). Nearly 140,000 wheat genes have been identified and mapped to the genome zipper maps of the 21 chromosomes of wheat for genetic mapping and plant breeding applications. Alien chromosomes, present as additions to the wheat genome, also can be isolated and sequenced, providing unlimited markers for alien gene introgression (Tiwari et al. 2014).
Laying the Foundation for Analysis of Chromosome Structure: Chromosome Banding and In Situ Hybridization Methods
By the 1970s, many genes had been mapped on wheat chromosomes and arms in relation to the centromere but little was known about the chromosome structure, especially chromatin differentiation into biological meaningful heterochromatic and euchromatic regions along the chromosome length. In the early 1970s, Gill and Kimber (1974a, b) applied the C-banding technique to wheat and rye revealing remarkable heterochromatic bands that allowed unequivocal identification of individual chromosomes. Combining C-banding with aneuploid stocks, they constructed cytogenetic maps of the chromosome complements of wheat and rye. A nomenclature system was developed to describe the hundreds of chromosome regions into which the wheat genome was partitioned for mapping, and idiograms of the 21 wheat chromosomes were constructed (Gill et al. 1991; Fig. 7.3). The homoeologous chromosomes belonging to A, B, D, and R genomes showed little conservation of chromosome structure, revealing the fast-paced tempo of cereal chromosome evolution in the Triticeae tribe.
(Left) The banded karyotype of Chinese Spring wheat. Each chromosome was stained (from left to right) by N-banding, modified C-banding, and C-banding (whole and telosomic chromosomes). Because chromosome 1A, and 3D to 6D do not show any N-bands, they were not identified. (Right) Idiogram of banded chromosomes of Chinese Spring wheat. Band numbers are indicated on the left and FL positions on the right of each chromosome. Hatched bands are not numbered as they were not observed consistently (Originally published in Gill et al. (1991). © Canadian Science Publishing or its licensors)
Next, newly developed in situ hybridization methods were used to map DNA sequences on chromosomes on a glass slide (Rayburn and Gill 1985). Repetitive DNA mapped to heterochromatic regions was often genome-specific (Rayburn and Gill 1986). Recently, a robust method of mapping single-copy sequences has been applied to wheat (Danilova et al. 2012). A set of 60 full-length cDNA clones for single-copy FISH was developed to rapidly determine the homoeology of any Triticeae chromosome with wheat chromosomes (Danilova et al. 2014; Fig. 7.4). This set also is very useful for detecting chromosomal rearrangements that may have occurred during Triticeae evolution.
Idiogram of wheat chromosomes where heterochromatic bands that can be visualized with GAA oligonucleotide probes are in black, pAs1 bands are in green, and cDNA probe positions are shown as red dots. Names of probes that hybridize to more than one homoeologous group are highlighted in red (Originally published in Danilova et al. (2014))
Laying the Foundation for Wheat Chromosome Manipulation: An Integrated Approach for Alien Genetic Transfers
Two basic methods of chromosome manipulation for alien gene introgression have been practiced widely in wheat. Sears (1956) used irradiation to transfer a leaf rust resistance gene from Aegilops umbellulata Zhuk. into wheat. Riley et al. (1968) interfered with the Ph1 gene effect (Riley and Chapman 1958) and used induced-homoeologous pairing to transfer yellow rust resistance from Ae. comosa Sm. in Sibth. & Sm. into wheat. Later, Sears (1977) produced a mutant at the Ph1 locus and also used a nullisomic 5B chromosome for induced homoeologous pairing and alien gene transfer (Sears 1973).
Chromosome banding and in situ hybridization greatly facilitated the characterization of the alien transfers in terms of the identity of the alien and wheat chromosomes involved in the translocations, the breakpoints of wheat–alien translocations, and the amount of the alien chromatin transferred (Friebe et al. 1991; Mukai et al. 1993; for review see Friebe et al. 1996). Qi et al. (2007) presented an integrated chromosome manipulation protocol utilizing modern cytological and molecular methods that has been widely used in many alien transfer experiments.
Looking to the Future
As the wheat genome is sequenced, it will be important to integrate DNA sequences with chromosome features such as heterochromatin and euchromatin, telomeres, and centromeres to gain insight into chromosome structure, function, and evolution, and, especially, how chromatin structure may control gene expression. The Triticeae tribe contains several 100 species, and only a few have been characterized by modern methods. Ideograms should be developed for all Triticeae chromosomes similar to the wheat chromosome idiogram (Fig. 7.3), including the mapping of single-copy FISH markers for elucidating syntenic relationships (Fig. 7.4). We also need to produce amphiploids, addition, substitution, and compensating Robertsonian translocations involving complete genomes of Triticeae species added to wheat. For accomplishing such a task, the IWGS provides a great platform for coordination of research and sharing of results.
References
Danilova TV, Friebe B, Gill BS (2012) Single-copy gene fluorescence in situ hybridization and genome analysis: Acc-2 loci mark evolutionary chromosomal rearrangements in wheat. Chromosoma 121:597–611
Danilova TV, Friebe B, Gill BS (2014) Development of a wheat single gene FISH map for analyzing homoeologous relationship and chromosomal rearrangements within the Triticeae. Theor Appl Genet 127:715–730
Endo TR, Gill BS (1996) The deletion stocks of common wheat. J Hered 87:295–307
Faris JD, Friebe B, Gill BS (2002) Wheat genomics: exploring the polyploid model. Curr Genomics 3:577–591
Friebe B, Mukai Y, Dhaliwal HS et al (1991) Identification of alien chromatin specifying resistance to wheat streak mosaic virus and greenbug in wheat germ plasm. Theor Appl Genet 81:381–389
Friebe B, Jiang J, Raupp WJ et al (1996) Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91:59–87
Gill BS, Chen PD (1987) Role of cytoplasm-specific introgression and evolution of the polyploid wheats. Proc Natl Acad Sci U S A 84:6800–6804
Gill BS, Kimber G (1974a) The Giemsa C-banded karyotype of rye. Proc Natl Acad Sci U S A 71:1247–1249
Gill BS, Kimber G (1974b) Giemsa C-banding and the evolution of wheat. Proc Natl Acad Sci U S A 71:4086–4090
Gill BS, Raupp WJ (1987) Direct genetic transfers from Aegilops squarrosa L. to hexaploid wheat. Crop Sci 27:445–450
Gill BS, Friebe B, Endo TR (1991) Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome 34:830–839
Gill BS, Friebe B, Raupp WJ et al (2006) Wheat Genetics Resource Center: the first 25 years. Adv Agron 85:73–135
Kihara H (1919) Uber cytologische studien bei einigen Getreidearten. I. Species-Bastarde des Weizens und Weizenroggen-Bastarde. Bot Mag Tokyo 32:17–38
Kihara H (1924) Cytologische und genetische Studien bei wichtigen Getreidearten mit besonderer Rucksicht auf das verhalten der Chromosomen und die Sterilitat in den Bastarden. Mem Coll Sci Kyoto Imp Univ B 1:1–200
Kihara H (1944) Discovery of the DD-analyzer, one of the analyzers of Triticum vulgare. Agric Hort (Tokyo) 19:889–890
Kihara H (1954) Considerations on the evolution of Aegilops species based on the analyzer-method. Cytologia 19:339–357
Law CN (1966) Location of genetic factors affecting quantitative character in wheat. Genetics 53:487–498
McFadden ES (1930) A successful transfer of emmer characters to vulgare wheat. J Am Soc Agron 22:1020–1034
McFadden ES, Sears ER (1944) The artificial synthesis of Triticum spelta. Rec Genet Soc Am 13:26–27
McFadden ES, Sears ER (1946) The origin of Triticum spelta and its free threshing hexaploid relatives. J Hered 37(81–89):107–116
Mukai Y, Friebe B, Hatchett JH et al (1993) Molecular cytogenetic analysis of radiation-induced wheat-rye terminal and intercalary chromosomal translocations and the detection of rye chromatin specifying resistance to Hessian fly. Chromosoma 102:88–95
Naranjo T, Rocca A, Goicoechea PG et al (1987) Arm homoeology of wheat and rye chromosomes. Genome 29:873–882
O’Mara JG (1940) Cytogenetic studies on triticale. 1. A method for determining the effects of individual Secale chromosomes on Triticum. Genetics 25:410–418
Qi LL, Echalier B, Chao S et al (2004) A chromosome bin map of 10,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168:701–712
Qi LL, Friebe B, Zhang P et al (2007) Homoeologous recombination, chromosome engineering and crop improvement. Chromosom Res 15:3–19
Rayburn AL, Gill BS (1985) Use of biotin-labeled probes to map specific DNA sequences on wheat chromosomes. J Hered 76:78–81
Rayburn AL, Gill BS (1986) Isolation of a D-genome specific repeated DNA sequence from Aegilops squarrosa. Plant Mol Biol Report 4:102–109
Riley R, Chapman V (1958) Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 182:713–715
Riley R, Chapman V, Johnson R (1968) The incorporation of alien disease resistance in wheat by genetic interference with the regulation of meiotic chromosome synapsis. Genet Res Camb 12:199–219
Sakamura T (1918) Kurze Mitteilung über die chromosomenzahlen und die Verwandschaftserhaltnisse der Triticum-Arten. Bot Mag Tokyo 32:151–154
Sax K (1922) Sterility in wheat hybrids. II. Chromosome behavior in partially sterile hybrids. Genetics 7:513–552
Sears ER (1939) Cytogenetic studies with polyploid species of wheat. I. Chromosomal aberrations in the progeny of a haploid of Triticum vulgare. Genetics 24:509–523
Sears ER (1954) Aneuploids of common wheat. Res Bull Mo Agric Exp Stn 572:1–59
Sears ER (1956) Transfer of leaf-rust resistance from Aegilops umbellulata to wheat. Brookhaven Symp Biol 9:1–22
Sears ER (1966) Nullisomic-tetrasomic combinations in hexaploid wheat. In: Lewis DR (ed) Chromosome manipulations and plant genetics. Oliver and Boyd, London, pp 29–47
Sears ER (1973) Agropyron-wheat transfers induced by homoeologous pairing. In: Sears ER, Sears LMS (eds) Proceedings of the fourth international wheat genetics symposium, Columbia. Missouri Agricultural Experiment Station, Columbia, pp 191–199
Sears ER (1977) An induced mutant with homoeologous pairing in common wheat. Can J Genet Cytol 19:585–593
Sears ER, Sears LMS (1978) The telocentric chromosomes of common wheat. In: Ramanujam S (ed) Proceedings of the fifth international wheat genetics symposium, New Delhi, pp 389–407
The International Wheat Genome Sequencing Consortium (IWGSC) (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:285–287
Tiwari VK, Wang S, Sehgal S et al (2014) SNP discovery for mapping alien introgressions in wheat. BMC Genomics 15:273
Acknowledgments
Contribution no. 15-169-A from the Kansas Agricultural Experiment Station. Research supported by the National Science Foundation and the Wheat Genetics Resource Center Industry/University Cooperative Research Center contract number IIP-1338897.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is distributed under the terms of the Creative Commons Attribution Noncommercial License, which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Copyright information
© 2015 The Author(s)
About this paper
Cite this paper
Gill, B.S. (2015). Wheat Chromosome Analysis. In: Ogihara, Y., Takumi, S., Handa, H. (eds) Advances in Wheat Genetics: From Genome to Field. Springer, Tokyo. https://doi.org/10.1007/978-4-431-55675-6_7
Download citation
DOI: https://doi.org/10.1007/978-4-431-55675-6_7
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-55674-9
Online ISBN: 978-4-431-55675-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)