Abstract
RNA interference (RNAi)-based gene silencing has become one of the most successful strategies in not only identifying gene function but also in improving agronomical traits of crops by silencing genes of different pathogens/pests and also plant genes for improvement of desired trait. The conserved nature of RNAi pathway across different organisms increases its applicability in various basic and applied fields. Here we attempt to summarize the knowledge generated on the fundamental mechanisms of RNAi over the years, with emphasis on insects and plant-parasitic nematodes (PPNs). This chapter also reviews the rich history of RNAi research, gene regulation by small RNAs across different organisms, and application potential of RNAi for generating transgenic plants resistant to major pests. But, there are some limitations too which restrict wider applications of this technology to its full potential. Further refinement of this technology in terms of resolving these shortcomings constitutes one of the thrust areas in present RNAi research. Nevertheless, its application especially in breeding agricultural crops resistant against biotic stresses will certainly offer the possible solutions for some of the breeding objectives which are otherwise unattainable.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
4.1 Introduction
RNA interference (RNAi) is an invaluable technology for unraveling gene function in the area of functional genomics. It has been utilized in basic research ranging from functional studies to gene knockdown in plants and vertebrates and to suppression of cancer and viral diseases in medicine. Moreover, from application point of view, it is being used extensively for trait modification by selective inhibition of gene expression universally across the organisms. In agriculture, RNAi has been extensively employed particularly for imparting resistance against biotic stresses including insects, bacteria, nematodes, fungal infection, and viruses (Tan and Yin 2004; Yanagihara et al. 2006; Good and Stach 2011; Banerjee et al. 2017; Majumdar et al. 2017; Zhang et al. 2017). This chapter focuses on how RNAi has been extensively used in managing various biotic stresses which constitute serious impediments to crop productivity. Damage due to insects, fungus, parasitic weeds, and plant-parasitic nematodes is a major biotic constraint causing significant yield losses in agriculture year-round.
4.2 History of RNAi
The basic concept involves a double-stranded RNA (dsRNA) molecule which potentially silences the gene with complementary sequences post-transcriptionally. RNAi phenomenon was first discovered in a free-living nematode, Caenorhabditis elegans (Fire et al. 1998). They coined the term “RNAi” for describing effective silencing of gene expression by exogenously supplied sense and antisense RNAs in the model nematode, Caenorhabditis elegans. This phenomenon, conserved among eukaryotes, was described as post-transcriptional gene silencing (PTGS) (Carthew and Sontheimer 2009; Berezikov 2011). Historically the roots of this exciting development can be traced back to 1990 when chsA gene was overexpressed in transgenic petunia plants and the silencing of endogenous as well as transgene of chalcone synthase in the transgenic plants was observed (Napoli et al. 1990). Loss of endogenous as well as transgene-derived mRNAs was described as co-suppression, a term formulated by Napoli. Soon, importance of this technology was well understood by the scientific community, and since then, phenomenal growth in this technology has taken place. In fungi, this mechanism of PTGS is known as quelling (Agrawal et al. 2003). In nature, viruses mediate PTGS in plants, and the effect is amplified in cytoplasm or in the nucleus.
4.3 Biogenesis and Mechanism of RNAi Pathway
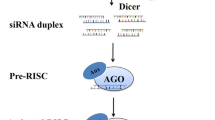
The major small noncoding RNAs (ncRNAs) include microRNAs (miRNAs), small interfering RNAs (siRNAs), and PIWI-interacting RNAs (piRNAs) which are all involved in downregulation of gene expression (Aalto and Pasquinelli 2012). Each class of small RNA is unique in its biogenesis and mechanism of action, but there are a few similarities too. Both miRNAs and siRNAs are processed from larger dsRNAs through cleavage by Dicer (a ribonuclease III enzyme). Both are associated with Argonaute proteins (AGO) (Ketting 2011) forming RNA-induced silencing complex (RISC). RISC basically is an Argonaute protein bound to a single strand of noncoding RNA. Varied ribonucleoprotein complexes arise due to several ncRNAs and Argonautes involved in formation of RISC (Darrington et al. 2017).
The RNAi-mediated gene silencing occurs basically in three stages (Siomi and Siomi 2009). First one involves processing of long dsRNA into small dsRNA by ribonuclease III; in the second stage, unwinding of these small RNAs leads to formation of one guide strand, which is loaded into the RISC, whereas the other strand known as passenger strand gets degraded. Finally, the RISC, directed by the guide strand, locates mRNAs containing sequences complementary to the guide, binds to these sequences, and either degrades the mRNA or blocks its translation (Winter et al. 2009). The mechanism of RNAi is emerging with all its complexity, but with clarity, as more and more players involved in the interference are getting identified and characterized.
The involvement of siRNA molecules as important intermediates of the RNAi process became evident through independent investigations carried out by researchers around the world. The first report of accumulation of siRNAs was confirmed by Hamilton and Baulcombe (1999) while studying tomato lines transformed with 1-aminocyclopropane-1-carboxyl oxidase (ACO) and later in Drosophila syncytial blastoderm embryo (Tuschl et al. 1999). Two other independent studies experimentally exhibited the 21–23 nucleotide small RNAs as intermediates for degradation of mRNA (Zamore et al. 2000; Elbashir et al. 2001). But how these small RNA molecules are excised from their precursor was yet to be discovered. As the role of RNase III enzymes had been recognized as dsRNA nucleases already, the RNase III domain-containing proteins were searched as one of the factors in siRNA biogenesis. Recently only, different experimental studies revealed the involvement of RNA-processing enzymes in chopping off the dsRNAs into siRNA molecules. One of the crucial enzymes, Dicer, was identified in Drosophila, by browsing its genome for the proteins dedicated for functioning like RNase III endonuclease activity (Bernstein et al. 2001). In another study, Dicer protein in C. elegans (a bidentate nuclease) was characterized revealing its functional role in small RNA regulatory pathways (Ketting et al. 2001). It was also deduced to be the ortholog of Drosophila DCR-1 protein. Ketting et al. (2001) in this study also showed the requirement of ATP for regulating the rate of siRNA synthesis. In yet another experiment reduction in ATP levels by 5000-fold in Drosophila revealed a decrease in the rate of siRNA production (Nykanen et al. 2001). It is now believed that Dicer acts as a complex of proteins with domains for dsRNA binding at its C terminus which are separable from motifs like helicase and PAZ. It was experimentally found to co-localize with an endoplasmic reticulum protein, calreticulin (Caudy et al. 2002). However, the role of ATP in the biogenesis of siRNA is abstruse due to its varied functions among different Dicer proteins in different organisms. An imperative involvement of ATPase in siRNA production was exhibited by Drosophila Dicer-2 and C. elegans Dcr-1 (Tomari and Zamore 2005) in contrast to human Dicer wherein an ATPase-defective mutant showed regular processing (Carthew and Sontheimer 2009). A comprehensive biochemical, molecular, genetic, and structural study revealed the presence of two main domains, namely, PAZ and RNaseIII, performing a crucial role in excising the siRNAs (Zhang et al. 2004; Macrae et al. 2006).
Once Dicer cuts off the dsRNA, synthesized siRNAs then enter the RISC complex. The double-stranded siRNAs act as a template for the RISC to recognize the complementary mRNA aided by Argonaute proteins. Agronaute proteins are required for the RISC assembly and have been biochemically characterized in Drosophila. Amplification of siRNAs has been reported in nematodes, fungus, plants and amoeba (Dykxhoorn et al. 2003). RNA-dependent RNA polymerase (RdRP) is proposed to be involved in augmenting the siRNA molecules on the basis of biochemical studies (Lipardi et al. 2001; Sijen et al. 2001). Sijen demonstrated the fundamental role of rrf1 gene having sequence homology to RdRP for the production of secondary siRNAs in C. elegans. In this study, the concept of transitive RNAi pathway induced by secondary siRNAs came into the picture. Thus, catalytic nature of RNAi was proposed.
4.4 RNAi in Insect Resistance
The direct loss in crop productivity due to damage by insect pest and the input-cost accrued in agrochemical based protection amount to billions of dollars every year worldwide. In spite of alarming environmental hazard directly due to residual toxicity of insecticides in food chain, the consumption of insecticides has been ever incremental. This is primarily due to resistance development in insect-pest population and lack of awareness among the farming community. The worldwide consumption of insecticide increases by almost 30% in every 4 years. Therefore, insect-pest management, preferably through an integrative approach and without indiscriminate use of insecticide, has become a most sought-after area in research planning worldwide. Millions of dollars were granted for researching on sustainable and low-cost alternate avenues of pest control strategies in five most important agricultural crops. Development of resistant cultivars in crops seems to be the most acclaimed alternative for minimizing the application of insecticides. Unfortunately, for most of the major crop- insect damage, either such resistant cultivars are not available or the resistance has been broken down. Further insight into such examples reveals that lack of resistance source maneuverable either through classical breeding or through transgenesis has been the major constraint.
Accessing unrelated gene pool through development of transgenics has emerged as the most potential avenue for overcoming this bottleneck. Success of Bacillus thuringiensis (Bt) toxin-mediated protection of a large number of crops has been celebrated widely and in fact demonstrated for the first time the potential of biotechnological means in developing genetic resistance. However, applicability of Bt-mediated protection is limited as many of the insect pests are not affected by Bt toxin, and also this technology has faced second-generation challenge of some major insect species developing resistance to Bt (Tabashnik 2008; Tabashnik et al. 2008). It has been realized that lack of useful insecticidal transgenes is the major limitation in transgenic-based engineering of genetic resistance. In contrary, through RNAi, any important gene can be precisely targeted to elicit lethality in the insect species. Use of RNAi has rapidly progressed for gene function analysis in various insect orders, including Diptera (Lum et al. 2003; Dietzl et al. 2007), Lepidoptera (Tian et al. 2009; Terenius et al. 2011), Coleoptera (Baum et al. 2007; Zhu et al. 2011; Bolognesi et al. 2012), and Hymenoptera (Nunes and Simoes 2009; Meer and Choi 2013; Zhao and Chen 2013).
4.5 RNAi Pathway in Insects
Like in plants, RNAi is primarily involved in antiviral defense mechanisms of insects as a part of its innate immunity. However, a number of studies indicate several branches of RNAi involved in endogenous gene regulation in addition to silencing of genetic elements of pathogen invaders and transposons (Van Rij and Berezikov 2009). Gene silencing through RNAi is systemic and transitive as originally described in C. elegans. A host-derived RNA-dependent RNA polymerase (RdRp) amplifies the RNAi post-elicitation by dsRNA. In contrast to nematodes, in insects, there is no definite proof of the presence of RdRp. In the absence of RdRp-mediated amplification of dsRNA in insects, the silencing is expected to be more localized. Therefore, elicitation of an effective silencing will require delivery of the dsRNA directly to the target cells and tissues in a continuous manner. The administered dsRNA enters the insect cells via siRNA pathway in which a complex consisting of the RNAase III enzyme (Dicer-2) and TRBP cuts the dsRNA into small 21–23 bpsiRNAs. The RISC bound to AGO recognizes the guide strands of the siRNAs. This complex then binds to complementary sequences of target RNAs which are eventually degraded.
Two types of RNAi pathway are known to occur in insects: cell-autonomous and non-cell-autonomous RNAi. Cell-autonomous RNAi is limited to the cells in which the dsRNA is administered or delivered. In contrary, when the silencing occurs in cells different from the cells delivered with or producing the dsRNA, it is called non-cell-autonomous RNAi. Depending on how the dsRNA is acquired by the cell, non-cell-autonomous RNAi can be grouped in two kinds: environmental RNAi and systemic RNAi. In environmental RNAi, dsRNA is absorbed by a cell from the surrounding environment. Therefore, this is seen in unicellular organisms or any cell lines when administered with dsRNA. Environmental RNAi does not necessarily result into systemic spread of the response. In multicellular organisms, silencing signal is transported from one cell to another by systemic RNAi.
In case of transgenic host-mediated delivery of dsRNA, the dsRNA is delivered into the gut lumen of insects. For eliciting effective RNAi, dsRNA must be taken up by gut cells from the gut lumen which is known as environmental RNAi. If the transcripts of target genes are prevalently expressed in tissues outside the gut cells, the systemic RNAi has to occur for spreading of silencing signal. However, there is no definite study on assessing systemic RNAi in insects.
4.6 RNAi in Plant-Parasitic Nematodes (PPNs)
Plant-parasitic nematodes (PPNs) are grouped on the basis of different type of lifestyles, i.e., sedentary, including root-knot nematode (RKN) and cyst nematodes, and migratory, including root-lesion nematodes. Sedentary endoparasites interact with the host through secretions which are vital cues for plant-nematode interactions. These secretory proteins are thus of major interest as targets for modulating the interaction. RNAi has been extensively used in functional genomics performed on C. elegans and opened up the possibility of deciphering the function of uncharacterized genes in other parasitic nematodes. Recent discoveries focused on unraveling the role of different components of RNAi in parasitic nematodes has eventually led to increasing our understanding of RNAi mechanism.
There are overwhelming reports on managing PPNs using RNAi. In nematodes, systemic RNAi can be observed resulting in a gene knockout that spreads throughout the organism. This is because RNA-dependent RNA polymerase (RdRP) is present in nematodes which interact with RISC and leads to production of new dsRNAs which are acted upon by Dicer enzymes and further produces new siRNAs (secondary siRNAs) in a well-coordinated amplification reaction. Therefore, the effect of dsRNA persists over development and also can be exported to neighboring cells thereby leading to silencing effect all over the organism (Daniel and John 2008). C. elegans displays systemic RNAi wherein the dsRNA/siRNAs entering from the environment can spread from one cell to another. Studies on identification of effectors of systemic RNAi revealed presence of protein SID-1 in C. elegans (Winston et al. 2002; Feinberg and Hunter 2003). Interestingly, M. incognita and M. hapla, along with other parasitic nematodes, despite exhibiting successful RNAi, were found deficient in SID-1 and other related proteins having a key role in dsRNA uptake and its spread. Several detailed comparative studies have postulated the presence of RNAi components in different PPNs and animal parasitic nematodes that were reported in C. elegans (Lendner et al. 2008; Dalzell et al. 2011; Haegeman et al. 2011). All these studies found rare proteins taking part in RNAi pathway. Seventy-seven orthologous effectors in C. elegans were searched in 13 nematode species, Ancylostoma caninum, Oesophagostomum dentatum, Ascaris suum, Brugia malayi, C. brenneri, C. briggsae, C. japonica, C. remanei, Haemonchus contortus, Meloidogyne hapla, M. incognita, Pristionchus pacificus, and Trichinella spiralis, using reciprocal BLAST followed by domain structure verification (Maule et al. 2011). It was concluded that effector deficiencies cannot, in any way, be associated with reduced susceptibility in parasitic nematodes. Surprisingly, minimum diversity was observed among these parasitic nematodes in most of the orthologous genes belonging to different functional groups (Table 4.1). Thus it was evident that all the species possess varied proteins from across the RNAi spectrum each with alternative proteins which are yet to be fully identified and characterized.
4.7 Mode of dsRNA Delivery
The efficacy of gene silencing substantially depends on the method of dsRNA uptake. In absence of systemic RNAi, gene silencing shall be limited to the cells that take up the dsRNA. Therefore, appropriate delivery system is pivotal (Terenius et al. 2011). Different delivery methods of dsRNA that have been used for successful RNAi in insects and nematodes include microinjection, feeding on either artificial diet (Table 4.2), and/or host-mediated delivery through transgenic plants (Fig. 4.1). Each of these methods has its own advantages and limitations.
4.7.1 Microinjection
Microinjection involves injection of dsRNA or siRNA directly into the body of an organism and has been demonstrated as one of the most successful delivery methods for RNAi to validate gene functions (Ober and Jockusch 2006). In this method, dsRNA is produced by in vitro transcription using T7 or Sp6 promoter sequences. It has been employed successfully for suppressing genes in both insects and nematodes.
4.7.1.1 In Insects
In D. melanogaster, microinjection has been successfully used for delivering dsRNAs for two genes, viz., frizzled and frizzled2, into embryos. The silencing resulted in defects in embryonic patterning that was similar to loss of wingless (wg) function. This was the first study proving the function of frizzle through dsRNA microinjection in an insect (Kennerdell and Carthew 1998). Since then, microinjection-based delivery has been used in several insect species. A comprehensive list of Hemipteran insects subjected to microinjection for studying RNAi is presented in Table 4.3. Direct injection of dsRNA into the insect body leads to higher efficiency of gene expression attenuation compared to other methods. Nevertheless, there are several limitations in microinjection delivery method. In vitro synthesis of dsRNA is skill intensive and costly. Additionally, recovery of the insects, especially smaller insects, from aftershock of microinjection, is relatively low. The significant aftershock is due to damage of cuticle leading to adverse immune responses in the insect (Roxstrom-Lindquist et al. 2004). Therefore, microinjection is rarely used in functional analysis of large number of genes from the point of view of insect-pest control. It is evident from Table 4.3 that in the microinjection, mediated delivery has been carried out mostly in the case of hemipteran insects.
4.7.1.2 In Nematodes
After injecting dsRNAs into the worms, progeny is counted and recorded for the mutant phenotypes. Usually after 24 h of injection, good RNAi effect is observed (Fire et al. 1998). In C. elegans, dsRNAs of genes like unc-22, unc-54, fem1, and hlh-1 were injected into the adult hermaphrodites, and the interference effect was observed. It was also proposed that in an antisense mechanism, interference of endogenous gene is due to the hybridization between the injected RNA and endogenous mRNA (Fire et al. 1998). It is a classical technique, and different target mRNAs can be used for injection simultaneously. However, microinjection has not been very successful in plant-parasitic nematodes in general and particularly in M. incognita. This is because of the small size of the infective stages and their inability to ingest fluid without host plant infection (Banerjee et al. 2017). In this process, although the range of dsRNA concentrations can be used, the success rate relies upon ample uptake or absorption by the worms (Hull and Timmons 2004).
4.7.2 Feeding on Artificial Diet
4.7.2.1 In Insects
dsRNA delivery through artificial diet has been the most popular method for delivering dsRNA into the insect gut especially for relatively smaller insects such as Hemipterans, which are sap-sucking. Several insect species of different taxa were studied for RNAi by the administration of dsRNA through artificial diet as presented in Table 4.3. Araujo et al. (2006) fed the blood-sucking Rhodnius prolixus with an artificial diet containing dsRNA of the nitrophorin2 (Np2) gene and found that the saliva of control R. prolixus prolonged plasma coagulation by approximately fourfold compared with the saliva of Np2-knockdown R. prolixus. Feeding A. pisum with an artificial diet supplemented with dsRNA of the A. pisum aquaporin 1 (ApAQP1) gene caused attenuated expression of the target gene, which resulted in an increased osmotic pressure of the hemolymph in this insect (Shakesby et al. 2009).
4.7.2.2 In Nematodes
In a nematode, feeding involves ingestion of bacteria expressing dsRNA of the target gene against which RNAi is employed. Timmons et al. (2001) developed engineered bacteria deficient for RNaseIII producing high levels of dsRNA segments of a specific gene. C. elegans feeding on these engineered bacteria showed RNAi effect leading to loss-of-function phenotypes for the target genes. One of the advantages of this method is that it can be conducted for stage-specific RNAi experiments as worms of any stage can be fed with dsRNA (Kamath et al. 2001; Ahringer 2006).
The feeding method has some major advantages over other methods of delivering dsRNA. These are as follows: (i) it is easy to perform; (ii) feeding dsRNA is less traumatic to the nymphs and juveniles than doing so via injections, the nymphs and juveniles remain healthier, and their mortality is comparatively lower (Shakesby et al. 2009); and (iii) perhaps most significantly, delivering dsRNAs in early stages of insects and nematodes is convenient by this method as compared to microinjection which needs special equipment and often causes high rate of mortality due to art effect. However, there are some challenges, viz., low efficiency of this method and requirement of large quantities of dsRNA, which need to be addressed. Moreover, a detailed study in understanding the mechanism of dsRNA delivery by ingestion for inhibiting gene expression is yet to be carried out.
4.7.2.3 Soaking Method for dsRNA Delivery in Nematodes
This method involves soaking of nematodes in concentrated dsRNA solution and subsequently scoring of worms or their progeny for phenotypes. RNAi by soaking is useful for treating a moderately large number of animals (e.g., 10–100). RNAi through soaking method was first employed in C. elegans as a tool for converting its genome sequence information into functional information (Tabara et al. 1998). Apart from C. elegans, silencing of genes in plant-parasitic nematodes (PPN) through soaking technique has been popularly used but with minor modifications. Other techniques like feeding and microinjection possess some limitations with respect to PPNs. In microinjection, successful recovery of injected juveniles is difficult and PPN juveniles do not take up dsRNA orally easily from the solutions. This was overcome by Urwin et al. (2002) by inducing oral uptake of dsRNA using octopamine, a neuroactive compound by cyst nematodes Heterodera glycines and Globodera pallida. This marked a revolution in imparting RNAi-mediated resistance in cyst and root-knot parasitic nematodes.
Since then many reports on successfully governing the nematode growth utilizing RNAi approach came into the picture. In later studies, compounds like resorcinol and serotonin were used for successful uptake of dsRNA in M. incognita (Rosso et al. 2005; Huang et al. 2006). Apart from neuroactive compounds, fluorescein isothiocyanate (FITC) as a marker for observing dsRNA uptake and as a mean of selecting affected individuals was used in many studies (Urwin et al. 2002; Rosso et al. 2005). Intestinal gene cysteine proteinase was suppressed through the soaking method in G. pallida, H. glycines, and M. incognita (Nakai and Horton 1999; Schmidt et al. 1999). Gene silencing by RNAi soaking has led to various abnormalities in processes like nematode hatching and molting and even resulted in reduced reproduction rates. Many genes, namely, chitin synthase, neuropeptides, msp, c-type lectin, and aminopeptidases, were targeted (Kennerdell and Carthew 1998; Schmidt et al. 1999; Dernburg and Karpen 2002; Ischizuka et al. 2002). But the efficiency and duration of the silencing effect were assessed for M. incognita calreticulin (Mi-crt) and polygalacturonase (Mi-pg-1) (Rosso et al. 2005). Other genes targeted by this approach are cellulases, pectate lyase, chorismate mutase, and glutathione-S transferase (Anandalakshmi et al. 1998; Cogoni and Macino 2000; Hammond et al. 2001; Matzke et al. 2001; Carmell et al. 2002). However, the silencing acquired by soaking in dsRNA solutions is often transient as duration of soaking and the concentration of dsRNAs affect the RNAi mechanism (Banerjee et al. 2017).
4.8 Resistance Via Transgenic Plants Expressing dsRNA
Another alternative method of dsRNA delivery is through host-delivered RNAi (HD-RNAi) where gene is silenced in target organism by the host plant. Since there is no synthesis of any gene product in HD-RNAi, it is likely to address the biosafety concerns more favorably.
4.8.1 In Insects
Genetic transformations of crop plants for expressing dsRNA homologous to important insect gene entail several advantages. It delivers the dsRNA to the target insect pest in a continuous fashion that leads to elicitation of RNAi throughout the life cycle of the insects. Host-mediated delivery of dsRNA was first demonstrated against two important agricultural pests, cotton bollworm, Helicoverpa armigera, and Western corn rootworm, Diabrotica virgifera (Baum et al. 2007; Mao et al. 2007). Transgenic rice was developed by delivering dsRNA targeting hexose transporter gene NIHT1, carboxypeptidase gene NIcar, and the trypsin-like serine protease gene NItry of Nilaparvata lugens. The study revealed reduced transcript levels of these three targeted genes in the insects that fed on these transgenic rice plants. However, insect lethality was not reported (Zha et al. 2011). Subsequently, several attempts have been made for attenuating key genes of the insects through transgenic host-mediated delivery of dsRNA as presented in Table 4.4. The gene construct for expression of the dsRNA essentially consists of 200–500 nucleotide tandem repeats of the target gene sequence under the control of a constitutive promoter. Such strategy also offers the scope of tissue specific expression of the dsRNA. For example, for targeting the phloem-feeding insect pests, phloem-specific expression of the dsRNA and their transport in phloem sieve elements would be more desirable. However, several attempts in this direction clearly indicated the effective level of protection would depend on targeting suitable target genes in addition to desired level of expression and delivery of intact dsRNA to the infesting insect pests (Price and Gatehouse 2008). Further understanding of the uptake process and elicitation of RNAi by dsRNA in insects will facilitate tailoring the gene expression cassette of dsRNA in order to achieve effective protection.
Mao et al. (2007) used RNAi-mediated approach to reduce insect’s ability to cope up when exposed to xenobiotic compounds, for example, gossypol. Transgenic cotton plants expressing a hairpin dsRNA targeting gossypol-inducible cytochrome P450 gene CYP6AE14 of H. armigera showed increased tolerance to the cotton bollworm, H. armigera (Mao et al. 2011), but were not lethal to the larvae. Interestingly, when a cysteine proteinase which is supposed to damage larval peritrophic matrix leading to higher accumulation of gossypol in the midgut was co-delivered, the tolerance was further enhanced (Mao et al. 2013). The similar strategy may be applicable for restoring insecticide sensitivity among resistant insect species (Bautista et al. 2009; Tanget al. 2012; Figueira-Mansur et al. 2013).
The host-mediated RNAi for controlling insect pest has been considered to be particularly important for phloem-sucking hemipteran insect pests, viz., aphids. In green peach aphid, plant-mediated RNAi of several target insect-specific genes such as salivary proteins MpC002, MpPIntO1, and MpPIntO2 and the gut-specific gene Rack-1 showed reduced fecundity (Table 4.3). In a similar study, stronger aphicidal activity of a hairpin RNA targeting V-ATPase E or the tubulin folding cofactor D (TBCD) was demonstrated (Guo et al. 2014). RNAi-mediated expression attenuation of a serine protease gene MySP in the green peach aphid, Myzus persicae, led to a remarkable decrease in their fecundity and parthenogeneticity (Bhatia et al. 2012). These studies on host-mediated delivery of dsRNA and elicitation of RNAi in infesting aphids demonstrated potential of RNAi approach for developing genetic resistance against aphids. Mao and Zeng (2014) reported reduced attack by aphids on transgenic tobacco plants expressing dsRNA against the gap gene hunchback, and reproduction rate of aphids was also retarded.
Interestingly, aphid nymphs parthenogenetically born from mothers reared on transgenic plants expressing dsRNA continued to show downregulation of the target gene even when transferred on normal plants. An assessment of RNAi effect over three generations of M. persicae revealed 60% reduction in aphid reproduction levels in transgenic Arabidopsis plants expressing dsMpC002 compared to 40% decline on transgenics expressing dsRack1 and dsMpPIntO2. Such transgenerational RNAi was found to last over seven generations in Sitobion avenae reared on transgenic barley plants expressing shp-dsRNA (Abdellatef et al. 2015). Such parental transmission of RNAi effect adds to potential of the strategy.
4.8.2 In Nematodes
RNAi mechanism partly occurs in the host itself and partly in nematodes feeding on the transgenic host plant expressing dsRNA for the target gene. The plant RNAi machinery produces siRNAs which are ingested by nematodes feeding upon these plants through stylet (Li et al. 2011). By far HD-RNAi is the most successful methodology for developing resistance against nematodes in important crops. This technique exploits the capability of PPNs of ingesting macromolecules from the host plants. Specifically, the method involves producing dsRNA construct and developing transformed plants by Agrobacterium-mediated transformation. For generating dsRNA, a part of the target gene is cloned in sense and antisense orientation separated by an intron or spacer region and expressed under a constitutive or tissue-specific promoter. Majority of researchers have adopted this time-consuming methodology and have successfully developed transgenics resistant against nematodes. Another new approach with rapid screening system has been developed involving hairy root method for transformation of crops like soybean, tomato, and sugar beet.
Genes involved in various vital processes are mostly targeted by this approach being categorized into effector genes (most targeted), house-keeping genes, developmental genes, and genes associated with mRNA metabolism. Two genes encoding integrase and splicing factor were suppressed in M. incognita using host-delivered RNAi. It was the first report eliciting RNAi in M. incognita by developing transgenic tobacco lines (Yadav et al. 2006). The lethality of these genes as RNAi targets was further reconfirmed by Kumar et al. (2017) in Arabidopsis by utilizing this approach against M. incognita. Effective silencing of 16D10 effector genes leads to 63–90% reduction in the infectivity of M. incognita in Arabidopsis (Huang et al. 2006). Since 16D10 is highly conserved in Meloidogyne species, resistance against three other major species was also developed (Li et al. 2011). M. chitwoodi also showed a reduction in the number of nematodes and eggs on silencing 16D10L gene via HD-RNAi approach in transgenic Arabidopsis and potato plants (Dinh et al. 2014a, b).
Cyst nematodes also exhibited gene suppression by this technique successfully. The suppression of four parasitism genes, ubiquitin-like (4G06), cellulose-binding protein (3B05), SKP1-like (8H07), and zinc finger protein (10A06), in Heterodera schachtii resulted in the reduction of females in RNAi transgenic Arabidopsis lines (Sindhu et al. 2009). Silencing of esophageal proteins in H. glycines leads to the reduction in reproduction (Bakhetia et al. 2007). In another study, successful suppression of major sperm protein of H. glycines resulted in 68% decrease in eggs per gram root tissue when infected on transgenic soybean plants (Steeves et al. 2006). Transgenic tobacco lines expressing dsRNAs of two neuropeptides, flp-14 and flp-18, showed 50–80% decline in the infection of M. incognita (Papolu et al. 2013). Other genes silenced using this methodology are Mj-Tisll, Rpn7, tyrosine phosphotase, mitochondria stress 70 protein precursor and neuropepetides against Meloidogyne spp(s) (Hamann et al. 1993; Lindbo et al. 1993; Depicker and Montagu 1997; Pasquinelli 2002; Lim et al. 2003; Valdes et al. 2003). Host-mediated RNAi strategy is more successful in root-knot (RKN) nematodes as compared to cyst nematodes (CN) owing to factors like more RNAi sensitivity and larger size exclusion limit of RKNs than in CNs (Li et al. 2011). Host-delivered RNAi appears to be the most successful technique in controlling nematode infection.
Identification of appropriate target genes based on preliminary diet-based bioassay and ensuring adequate in planta expression of the dsRNA in the transgenic host are pivotal requirements for effective host-mediated RNAi. However, further understanding of the mechanisms on dsRNA uptake by insect and nematodes will facilitate the tailoring of dsRNA expression in HD-RNAi.
4.9 dsRNA Uptake Mechanisms
The dsRNA uptake mechanism in insects is known to be achieved by either of the two pathways, viz., a protein-mediated pathway and via endocytic pathway. The major component of protein-mediated pathway is a multi-pass transmembrane protein known as systemic RNA interference deficient-1 (Sid-1) which exports the small interfering RNAs across neighboring cells (Bansal and Michel 2013). The second pathway is receptor-mediated pathway. In case of C. elegans, the endocytic pathway involves a Sid-2 gene localized in intestinal cells. It encodes a membrane protein and is thought to import dsRNA from the intestinal lumen which are then exported to other cells with the help of sid-1 channels (Winston et al. 2007; McEwan et al. 2012). Hence, Sid-1 and Sid-2 proteins must work in conjunction to achieve environmental RNAi. Sid-1 genes have been reported to be evolutionarily conserved among insects orders, but Sid-2 gene is absent in insects. Tribolium is considered as the model insect for studying systemic RNAi with presence of Sid-1 like proteins. However, the Sid-1 gene of Tribolium was found orthologous to Tag-130 gene of C. elegans and not Ce-Sid-1 gene interestingly, where Tag-130 has not been reported to be associated with systemic RNAi in nematodes (Tomoyasu et al. 2008). The presence of Sid-1-like channel proteins varies among different orders of insects. The involvement of Sid-1-like channel proteins in dsRNA uptake has been reported in brown plant hopper [BPH, Nilaparvata lugens (Xu et al. 2013)], the Colorado potato beetle [CPB, Leptinotarsa decemlineata (Cappelle et al. 2016)], and the red flour beetle Tribolium castaneum (Tomoyasu et al. 2008). In 2016, genes involved in RNAi pathway in insects were identified and classified. The study reveals absence of Sid-1/Tag-130 orthologs in Diptera order (Dowling et al. 2016). It was suggested that in Drosophila melanogaster, dsRNA uptake is mediated via endocytic pathway along with pattern recognition receptors (PRRs) based on a study by Ulvila et al. (2006). This study reports more than 90% reduction in the uptake of double-stranded RNA on silencing of these two receptors by RNAi technology. Most of the studies examining dsRNA uptake so far focused on either the endocytic pathway or Sid-1-like dependent system. However, a clear understanding of the roles of these pathways on dsRNA uptake across the insect species is still lacking. Nevertheless, insects belonging to another order have been reported to have both the Sid-1-like channel proteins and receptor-mediated endocytosis pathways playing a role in dsRNA uptake (Cappelle et al. 2016).
However, the dsRNA uptake mechanism in worms is quite different. The components involved in dsRNA uptake have been well studied in C. elegans, and presence of Sid-1 and Sid-2 genes along with other components like rsd-2, rsd-3, and rsd-6 has been well documented in the C. elegans genome. But surprisingly in a study, it was found these proteins were not evolutionary conserved (Dalzell et al. 2011). The dataset recognizes sid-1orthologs in two parasitic nematodes, viz., in Haemonchus contortus and Oesophagostomum dentatum only. The Sid-2 protein was not found to be present in other nematode species. Intriguingly, the plant-parasitic nematodes such as Meloidogyne and Globodera spp. despite the absence of Sid-1 and Sid-2 genes exhibit systemic RNAi when subjected to silencing technology indicating a presence of similar receptor-mediated endocytic process for dsRNA uptake as reported in insects (Dalzell et al. 2011). Though lot of information has been generated over past few years, a clear understanding on dsRNA uptake mechanism(s) in worms is still elusive
4.10 RNAi Resistance in Other Agricultural Pests
Other than insects and nematodes, there are agricultural pests belonging to phylum Arthropoda that affect the crop productivity worldwide, and RNAi-based strategy to control these pests has shown some success. These pests are fire ants, mites, locusts (order Orthoptera), and many more. Systemic RNAi has already been demonstrated in these pests via microinjection. On feeding the worker ants, Solenopsis invicta, with 1000 ppm dsRNA targeting PBAN/pyrokinin gene, increased mortality rate of the fourth instar larvae. Direct toxic effect was also observed even when the dsRNA concentration was reduced to 200 ppm (Zhao and Chen 2013). In spider mite, gene silencing and increased mortality rate was observed when 160 ppm of dsRNA, targeting several genes, was employed via permeated leaf disc assay (Kwon et al. 2013). In another mite, Varroa destructor, an ectoparasite of the honey bee, Apis mellifera, both the delivery methods of dsRNA, i.e., by immersing mites in a dsRNA solution or by host-mediated RNAi, wherein dsRNA was fed to the honey bees and eventually delivered to mites, were found to attenuate the target gene expression through environmental RNAi (Campbell et al. 2010; Garbian et al. 2012).
Interestingly, locust species displayed systemic RNAi response but were refractory to environmental RNAi. Even a considerate concentration of 15 pg of dsRNA per mg body mass (~10 ng/insect) was enough to silence a gene in the desert locust, Schistocerca gregaria (Wynant et al. 2012). In case of Tribolium castaneum, the systemic response continued to increase over time in a dose-dependent manner and furthermore led to mortality 7 days postinjection. A similar dose-dependent response was also exhibited by the migratory locust, Locusta migratoria, leading to target gene suppression and lethality, but was unresponsive to environmental RNAi (Luo et al. 2013).
4.11 RNAi for Fungus Resistance
Fungi are classified as a separate eukaryotic kingdom from plants and animals. The vital RNAi components (RNA-dependent RNA polymerase (RdRP), Dicer, and Argonaute) have been found in different fungi indicating the presence of functional RNAi pathway (Dang et al. 2011). The RNAi phenomenon is termed as “quelling” in fungi which was first demonstrated in ascomycete Neurospora crassa (Romano and Macino 1992). Silencing of fungal genes by RNAi has shown to be desirable for many fungal species like Ascomycota, Basidiomycota, Zygomycota, and Phytophthora species (Nunes and Dean 2012). Several studies have been published reporting the successful use of host-induced gene silencing (HIGS) to control fungal diseases (Table 4.5) (Koch and Kogel 2014). Suppression of GUS transcripts in a GUS-expressing strain of Fusarium verticillioides (phytopathogenic filamentous fungi) while colonizing transgenic tobacco plants expressing GUS gene-interfering cassette was reported (Tinoco et al. 2010).
In vitro feeding of dsRNA complementary to three genes involved in ergosterol biosynthetic pathway, viz., CYP51A, CYP51B, and CYP51C, showed reduced growth of Fusarium graminearum (Koch et al. 2013). In wheat, mycotoxin-specific genes were silenced in F. graminearum and resulted in inhibition of virulence (McDonald et al. 2005). Fungal pathogenicity genes have shown to be an appropriate target for controlling fungal infection. A complete loss of pathogenicity was reported on targeting two of the host-selective ACT-toxin genes in the fungus Alternaria alternata (Miyamoto et al. 2008; Ajiro et al. 2010). Similar reports on silencing of pathogenicity gene or avirulent gene proved successful in inhibiting the fungal growth and development. In Magnaporthe oryzae, silencing of 37 genes involved in calcium signaling process adversely affected hyphal growth, sporulation, and pathogenicity (Nunes and Dean 2012). HIGS-mediated silencing of effector gene Avra10 showed a reduction in the number of haustoria in powdery mildew-susceptible barley cultivar (Koch and Kogel 2014).
To date, there are several successful reports of gene silencing in fungi with varied silencing efficiency. For instance, in Moniliophthora perniciosa, the silencing efficiency varied depending upon the targeted gene with reduction rates ranging from 18% to 97% in case of hydrophobin transcripts and 23% to 87% in peroxiredoxin transcripts (Santos et al. 2009), while when RNA hairpin precursor used to transform the Ascomycota Ophiostoma novo-ulmi, the expression of 6%, 22%, and 31% relative to the wild type was reported (Carneiro et al. 2010) in three transformants. Although usage of RNAi for managing fungus growth is nowadays a favored approach by researchers, RNAi silencing also leads to some off-target effects as observed by Lacroix and Spanu (2009) on silencing various genes in C. fulvum. These off-targets can be avoided by using specific silencing trigger sequence in RNAi vector, by tissue-specific and inducible silencing (Senthil-Kumar and Mysore 2011).
4.12 Barriers Limiting RNAi
The potential of RNAi technology for controlling various pests has been well documented over the past decade. However, there are many limitations which need to be taken care of for successful deployment of RNAi technology. There are several factors which need to be carefully looked into while designing RNAi experiments, including the off-target effects, dsRNA design, length and concentration of dsRNA, and many more. Therefore, to ensure a successful and effective RNAi-based silencing, these factors need to be balanced optimally. In case of insects, persistency of RNAi is a major problem due to which an optimum amount of dsRNA needs to be determined for an effective silencing. Interestingly, it is not true for every order of insect which is to be managed. For instance, about 60% (or lower) of gene knockdown was reported in certain recalcitrant insect species, while in coleopterans, 90% knockdown of gene was successfully achieved ensuing a long-lasting hereditary (Baum et al. 2007; Huvenne and Smagghe 2010; Zhu et al. 2011; Bolognesi et al. 2012; Rangasamy and Siegfried 2012; Li et al. 2013). Not only in insects but in nematodes also barriers like off-target effects have been reported while performing RNAi technology based management approaches. Designing an effective siRNA sequence is a major limitation in RNAi technology-based silencing. The following are some major barriers.
4.12.1 Off-Target Effects
Off-target effects result from the knockdown of unintended genes other than the target gene. Therefore, one of the most important aspects is avoidance of nonspecific target effects. It is the sequence used that determines possible off-target effects in the target organism and also in other species. Other than sequence, off-target effects can arise due to wide range of siRNAs being produced from a single dsRNA which increases the chance of nontarget effects. There are many reports of off-target effects, for instance, in triatomid bug R. prolixus, two homologous nitroprin genes were silenced other than the targeted gene (Araujo et al. 2006). Thus, selecting a sequence for synthesizing dsRNA is a crucial and limiting step in RNAi technology.
4.12.2 The Design of dsRNA
Selection of target gene is the first step in decision-making process for successful induction of RNAi in an organism. The gene selected should have a crucial role in the concerned organism, and genes involved in parasitism or development are likely candidate genes fulfilling all such requirements. Moreover, it should be highly specific and not conserved across different genera (Danchin et al. 2013) especially in pollinators. Next stage is to choose a suitable target site from the selected target gene. It is necessary to ensure the designing of a species-specific dsRNA. For identifying potential target sites for eliciting effective RNAi, bioinformatic tools are available online. Specificity of the dsRNAs could be conferred by either targeting conserved domain or variable region depending on the candidate gene with the aim to minimize possibility of affecting any unintended genes or organisms. This is particularly important to ensure that dsRNAs targeting agricultural pests should not possess any overlapping similarity to the genes of beneficial pollinators. By targeting the UTR regions, even closely related homologous genes can be selectively silenced through RNAi as demonstrated in D. melanogaster, T. castaneum, A. pisum, and tobacco hornworms, Manduca sexta, with respect to vATPase gene (Whyard et al. 2009).The concept of dsRNAs being used as tailor-made pesticides is emerging wherein highly specific dsRNAs are employed against havoc-creating pests and are also eco-friendly to the environment.
4.12.3 Length and Concentration of dsRNA
In general, longer RNA molecules tend to have longer half-life and therefore may be considered desirable while designing dsRNAs. However, size of the dsRNA molecule could be a limiting factor toward efficient uptake by the organisms. In nematodes, 28–140 kDa dsRNA could be efficiently ingested by Meloidogyne species (Urwin et al. 1997; Li et al. 2007; Zhang et al. 2012), though the limit is not known for other pests. In red flour beetle, the length and concentration of dsRNA had profound effect on efficiency as well as persistence of the RNAi effect, for example, 60- and 30-bp dsRNAs induced 70 and 30% of gene knockdown, respectively (Miller et al. 2012). In the same study, it has been also shown that multiple dsRNAs, when injected together, led to competitive inhibition influencing the effectiveness of RNAi. In contrary, dsRNA longer than 200 nucleotides and likely to generate multiple siRNAs contribute efficient RNAi response (Andrade and Hunter 2016). Multiple siRNAs will help in overcoming the target resistance that may arise due to polymorphism in the target. However, more studies are warranted to understand unambiguously the effect of length and concentration of dsRNAs on the initial efficiency and persistence of the RNAi effect.
4.12.4 Screening of Target Genes
For realizing RNAi-mediated gene silencing as an applicable strategy of pest control in agriculture, it remains imperative to achieve significant mortality or growth arrest of the pest population. Therefore, any attenuation of the target gene must be indispensible for the pest organism. This in turn underlines the importance of identifying appropriate target gene for the target pest. Though most of the studies have used limited set of target genes reported earlier, more emphasis should be given on identification of novel candidate genes (Pitino et al. 2011; Zhu et al. 2011). The upcoming genomics and bioinformatics tools, like genome search (Bai et al. 2009), cDNA library (Mao et al. 2007; Baum et al. 2007), RNA-seq and digital gene expression tag profile (DGE-tag) (Wang et al. 2011), and RIT-seq (Alsford et al. 2011), have been used for identification of new target genes.
4.12.5 Persistence of the Silencing Effect
The persistence of silencing signal determines the effectiveness of RNAi. Studies on low persistence of silencing effect have been reported in A. pisum wherein silencing effect on aquaporin persisted for 5 days of delivery before subsiding (Shakesby et al. 2009) indicating transient nature of RNAi effect. Thus, continuous supply of dsRNA seems to be essential for effective RNAi. It lends support for the transgenic host-mediated expression of the dsRNA for persistent and effective silencing. Persistent RNAi will also be useful in manifesting desired effect on the target organism even in case of inefficient and partial downregulation of the target gene.
4.12.6 Life Stage of the Target Organism
Selecting a life stage for larger silencing effects is species dependent that is to be targeted. In most cases, younger stage is preferred despite the efficient handling of older stages. In plant-parasitic nematodes, selecting the pre-parasitic juvenile stage for delivering dsRNAs shows better silencing effect. Similar observation was reported in insects, for example, in case of R. prolixus, no silencing effect was observed after treating its fourth instars compared to 42% silencing when using second instars (Araujo et al. 2006).
4.12.7 Methods of Delivery and Uptake Mechanisms
Various methods of dsRNA delivery have been used across the organisms. Such methods include microinjection, feeding with bacteria expressing dsRNA, feeding through diet supplementation, and host-mediated ingestion. The efficiency of RNAi varies significantly among different organisms and when using different delivery methods. In insects, either microinjection or diet supplementation has been the method of choice, though the aftershock effect of microinjection remains a concern in many species. Microinjection-mediated direct delivery bypasses the exposure of the dsRNA molecule to the nucleases present in the digestive tract. However, for realizing true efficacy of the dsRNA, it is desired to deliver through oral delivery that mimics the host-mediated delivery through ingestion.
4.12.8 Nucleases and Viruses
Limited success in RNAi in some of the insects has been attributed to rapid degradation of dsRNA by saliva of the insects. The saliva of Lygus lineolaris was found to contain RNases which interact with plant material prior to ingestion (Allen and Walker 2012). Presence of nucleases in the saliva and viruses in the hemolymph of insects also limits the silencing efficiency by degrading dsRNAs (Thompson et al. 2012; Christensen et al. 2013).
4.13 Improving RNAi
4.13.1 Large Throughput Screening for Selection of Target Genes
An ample number of studies in insect orders of Coleoptera, Diptera, Lepidoptera, Hemiptera, and others comprising of several insect pests have shown that RNAi targeting insect genes can affect growth and development of insects, often leading to insect death (Tables 4.3 and 4.4). The kind of genes for which a relatively high RNAi efficiency could be achieved included genes encoding detoxification enzymes, metabolism and cytoskeleton structure, cell synthesis, nutrition, etc. Alternative pathways of many of these genes in insects as well as relative importance of a particular pathway in an insect species are not known with certainty. Therefore, use of RNAi as a strategy for pest control will require an essential step of target selection. If an indispensible gene has to be identified for an insect species, it will involve large throughput screening rather than going for homologous genes, effective for other insect species.
Chitin covers the exoskeleton of insect body, and the insect midgut lined by peritrophic membrane (PM) constitutes the major channel for absorption of nutrients as well as orally administered dsRNA. Therefore genes expressed and functioning in the insect midgut have been screened by many researchers (Wang and Granados 2001). For example, a chitinase gene (OnCht) and a chitin synthase gene (OnCHS2) were identified from gut-specific EST of European corn borer (Ostrinia nubilalis) (Khajuria et al. 2010). Chitin content of the PM is regulated by OnCht as demonstrated in feeding experiment with dsRNA- and RNAi-based suppression which led to reduced growth and development of European corn borer larvae (Khajuria et al. 2010). In a similar study, Mao et al. (2007) identified several gossypol inducible genes, including a putative P450 monooxygenase, CYP6AE14, from a midgut-specific cDNA library from fifth-instar larvae exposed to gossypol. Similarly, for screening targets for RNAi in coleopteran insects, a large number of cDNAs from the cDNA libraries of Western corn rootworm (Diabrotica virgifera virgifera) were in vitro transcribed and used in feeding-based bioassays (Baum et al. 2007).
A rapid method of cDNA screening was demonstrated by Wang et al. (2011) by combining Illumina’s RNA-seq and digital gene expression tag profile (DGE-tag) in Asian corn borer (ACB) (Ostrinia furnacalis). In addition to being a rapid and cost-effective method, this method allows monitoring expression of the genes throughout the insect body and thus broadening the base of target selection. Using Illumina parallel sequencing technology, abundance of >90,000 transcripts from trypanosome libraries was scored before and after induction of RNAi. The results led to constitution of non-redundant set of protein-coding sequences (CDS) comprising ∼7500 genes (Alsford et al. (2011). Thus these methods can derive core set of essential gene loci if genome sequence of the organism is known. RNAi-mediated attenuation of these core loci is most likely to significantly retard survival and fitness of the insect pests.
In recent years, several modifications and methods for effective delivery and uptake of dsRNA have been proposed. Such methods include chemical modifications of siRNA duplex delivery through nanoparticles and liposomes, sprayable RNAi-based products, root absorption and trunk injection, and bacteria- or virus-based delivery. A few of them with much potentiality have been described below.
4.13.2 Nanoparticles
Synthetic, nontoxic nanoparticles could be generated from natural as well as synthetic polymers. Nanoparticles offer ease of surface modifications and biodegradability in addition to more penetration ability, thus an effective vehicle for delivery of dsRNA (Vauthier et al. 2003; Herrero-Vanrell et al. 2005). In mosquito dsRNA encapsulated in polymer, chitosan was used to achieve RNAi (Zhang et al. 2010). The encapsulation process used the electrostatic forces between the negative charges of the RNA backbone and positive charges of the amino groups of chitosan. Zhang et al. (2015a, b) demonstrated effective knockdown of AgCHS1 and AgCHS2 in A. gambiae and A. aegypti (sema1a) during larval development by using chitosan nanoparticles. He et al. (2013) fed lepidopteran pest, Asian corn borer (Ostrinia furnacalis), with diet containing the mixture of fluorescent nanoparticle (FNP) and CHT10-dsRNA, naked CHT10-dsRNA, FNP and GFP-dsRNA, and GFP-dsRNA. RNAi-mediated gene silencing occurred only in the larvae fed on the diet containing the mixture of FNP and CHT10-dsRNA leading to retarded growth and eventually death of the larvae.
4.13.3 Liposomes
Liposome vesicles composed of nontoxic natural lipids are already being used in drug delivery. Liposomes can cross the cell membrane effectively and deliver the exogenous molecules. Whyard et al. (2009) used cationic liposomes for encapsulating and delivering dsRNA targeting 3′-UTR of the g-tubulin gene in four different species of Drosophila (D. melanogaster, D. sechellia, D. yakuba, and D. pseudoobscura) and demonstrated mortality of the insects only in case of encapsulated dsRNA. In Drosophila, presence of sid1 homologues has never been confirmed, and the uptake of dsRNA is likely to be by receptor-mediated endocytosis (Ulvila et al. 2006). Higher efficiency of RNAi in case of liposome-mediated delivery in certain cases could be attributed to the fact that it bypasses the gut nucleases which reduces the efficacy of orally delivered dsRNA.
4.13.4 Chemical Modifications
Chemical modifications are known to increase the stability of RNA molecules. In case of siRNA also such modifications have been proposed to improve half-life and pharmacokinetic properties of the siRNA duplexes, target-binding affinity, and delivery (Kurreck 2003; Manoharan 2003; Dorsett and Tuschl 2004). Interestingly a couple of examples have demonstrated that such modifications may increase the specificity of dsRNA. For example, methylation at 2′-position of the ribosyl ring of the second base of the siRNA could decrease off-target effects (Jackson et al. 2003), siRNA duplex with 3′-overhangs at each end was more effective in gene silencing compared to blunt-ended duplex (Elbashir et al. 2001), and addition of 3′-TT overhangs (the “Tuschl design”) on both strands of duplex siRNA has been preferred in many cases. A few other designs, for instance, siRNAs without 3′-overhangs and single 3′-overhang structures in the guide strand, have been active in gene silencing (Czauderna et al. 2003; Lorenz et al. 2004).
4.14 Future Perspective and Conclusion
Despite few limitations, the applicability of RNAi in improving crop resistance especially against biotic stresses is expected to be the most reliable and significant approach in the future as evident from a plethora of studies. Certain products based on RNAi-mediated resistance such as Monsanto’s SmartStax Pro, for control of Western corn earworm, and DuPont Pioneer’s Plenish high oleic acid soybean (Majumdar et al. 2017) are likely to be commercialized soon. However, efficacy of these plants remains to be proven in actual field situations. Diverse classes of biotic factors, affecting crop production worldwide, have shown varied levels of susceptibility toward RNAi, which warrants need for modified and improved versions of dsRNA delivery methods. The better understanding of host-pest interaction and the genetic basis of parasitism are likely to generate more potential target genes for effective HD-RNAi. CRISPR/Cas system has come up as a powerful technique in creating knockout mutants to unravel complex mechanism of parasitism and thus paves the way for identification of the key pest genes. Transplastomic expression of dsRNA in the plants would be a further improvement for achieving higher expression. Applying dsRNA through methods with low environmental risks, for instance, irrigation water, root drench, or trunk injection, would obviate the need for genetic transformation. These methods result in localized application along with rapid breakdown of dsRNA and therefore likely to be more acceptable from a biosafety point of view (Joga et al. 2016). Successful demonstration of using layered double hydroxide clay nanosheets for topical application of dsRNA against viruses (Mitter et al. 2017) opens up possibilities of applying dsRNA like any other protective agrochemicals.
To conclude, RNAi has emerged as one of the most potential control mechanisms for pests like insects, nematodes, fungus, etc. Although still a lot remains to be explored and understood about the molecular process of RNAi in plants and their pests, the present available knowledge and the studies reviewed in this chapter have proved RNAi technology as an important tool in identifying gene functions and targeting vital genes for controlling pest development. RNAi-mediated loss-of-function phenotypes not only determine functions of unknown genes but also lead to identification of new specific targets for managing pest or improving agricultural traits. But understanding RNAi mechanism is of utmost importance as RNAi machinery varies from genus to genus. There are several shortcomings that need to be addressed, for instance, persistence of silencing effects, off-target effects of silencing, etc. Not only this, the biosafety, risk assessment, and government regulations related to commercialization of RNAi-based transgenics still have to be developed. The revelation of RNAi technology has revolutionized the area of research in biotechnology. Not only in pest management, the wide range of RNAi application includes modification of agronomic traits, eliminating mycotoxin contamination, improving nutritional value of crops, etc. It is also proving its worth in RNAi-based therapeutics research for human welfare. In toto, this technology is a potential boon in the arsenal of the scientific community to address the challenges associated with climatic changes, burgeoning population, and sustainability of human race.
References
Aalto AP, Pasquinelli AE (2012) Small non-coding RNAs mount a silent revolution in gene expression. Curr Opin Cell Biol 24:333–340
Abdellatef E, Will T, Koch A et al (2015) Silencing the expression of the salivary sheath protein causes transgenerational feeding suppression in the aphid Sitobion avenae. Plant Biotechnol J B13:849–857
Agrawal N, Dasaradhi PVN, Mohmmed A et al (2003) RNA interference: biology, mechanism and applications. Microbiol Mol Biol Rev 67:657–685
Ahringer J (ed) (2006) The C elegans research community. Reverse genetics J. http://www.wormbook.org
Ajiro N, Miyamoto Y, Masunaka A et al (2010) Role of the host-selective ACT-toxin synthesis gene ACTTS2 encoding an enoylreductase in pathogenicity of the tangerine pathotype of Alternaria alternata. Phytopathology 100:120–126
Allen ML, Walker WB (2012) Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J Insect Physiol 58:391–396
Alsford S, Turner DJ, Obado SO et al (2011) High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res 21:915–924. https://doi.org/10.1101/gr.115089.110
Anandalakshmi R, Pruss GJ, Ge X et al (1998) A viral suppressor of gene silencing in plants. Proc Natl Acad Sci U S A 95:13079–13084
Andrade CE, Hunter WB (2016) RNA interference– natural gene-based technology for highly specific pest control (HiSPeC). In: Abdurakhmonov IY (ed) RNA interference. InTech, Croatia, pp 391–409
Antonino JD, Coelho R, Lourenço T et al (2013) Knocking- down Meloidogyne incognita proteases by plant-delivered dsRNA has negative pleiotropic effect on nematode vigor. PLoS One 8:e85364. https://doi.org/10.1371/journal.pone.0085364
Araujo RN, Santos A, Pinto FS et al (2006) RNA interference of the salivary gland nitrophorin 2 in the triatomine bug Rhodnius prolixus (Hemiptera: Reduviidae) by dsRNA ingestion or injection. Insect Biochem Mol Biol 36:683–693
Bai J, Sepp KJ, Perrimon N (2009) Culture of Drosophila primary cells dissociated from gastrula embryos and their use in RNAi screening. Nat Protoc 4:1502–1512
Bakhetia M, Charlton W, Atkinson HJ et al (2005) RNA interference of dual oxidase in the plant nematode Meloidogyne incognita. Mol Plant Microbe Interact 18:1099–1106. https://doi.org/10.1094/MPMI-18-1099
Bakhetia M, Urwin PE, Atkinson HJ (2007) qPCR analysis and RNAi define pharyngeal gland cell-expressed genes of Heterodera glycines required for initial interactions with the host. Mol Plant Microbe Interact 20:306–312. https://doi.org/10.1094/mpmi-20-3-0306
Banerjee S, Banerjee A, Gill SS et al (2017) RNA interference: a novel source of resistance to combat plant parasitic nematodes. Front Plant Sci 8:834. https://doi.org/10.3389/fpls.2017.00834
Bansal R, Michel AP (2013) Core RNAi machinery and Sid1, a component for systemic RNAi, in the Hemipteran insect, Aphis glycines. Int J Mol Sci 14:3786–3801. https://doi.org/10.3390/ijms14023786
Baum JA, Bogaert T, Clinton W et al (2007) Control of coleopteran insect pests through RNA interference. Nat Biotechnol 25:1322–1326. https://doi.org/10.1038/nbt1359
Bautista MA, Miyata T, et a MK (2009) RNA interference-mediated knockdown of a cytochrome P450, CYP6BG1, from the diamondback moth, Plutella xylostella , reduces larval resistance to permethrin. Insect Biochem Mol Biol 39:38–46
Berezikov E (2011) Evolution of microRNA diversity and regulation in animals. Nat Rev Genet 12(12):846–860
Bernstein E, Caudy AA, Hammond SM et al (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363–366
Bhatia V, Bhattacharya R, Uniyal PL et al (2012) Host generated siRNAs attenuate expression of serine protease gene in Myzus persicae. PLoS One 7(10):e46343. https://doi.org/10.1371/journal.pone.0046343
Bolognesi R, Ramaseshadri P, Anderson J et al (2012) Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoSONE 7:e47534. https://doi.org/10.1371/journal.pone.0047534
Bonning BC, Chougule NP (2014) Delivery of intrahemocoelic peptides for insect pest management. Trends Biotechnol 32:91–98
Campbell ME, Budge GE, Bowman AS (2010) Gene-knockdown in the honey bee mite Varroa destructor by a non-invasive approach: studies on a glutathione S-transferase. Parasit Vectors 3:73. https://doi.org/10.1186/1756-3305-3-73
Cappelle K, de Oliveira CFR, Eynde BV et al (2016) The involvement of clathrin-mediated endocytosis and two Sid-1-like transmembrane proteins in double-stranded RNA uptake in the Colorado potato beetle midgut. Insect Mol Biol 25:315–323
Carmell MA, Xuan Z, Zhang MQ et al (2002) The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 16:2733–2742
Carneiro JS, Bastide PY, Chabot M et al (2010) Suppression of polygalacturonase gene expression in the phytopathogenic fungus Ophiostoma novo-ulmi by RNA interference. Fungal Genet Biol 47:399–405
Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655
Caudy AA, Myers M, Hannon GJ et al (2002) Fragile X-related protein and VIG associate with RNA interference machinery. Genes Dev 16:2491–2496
Chen Q, Rehman S, Smant G et al (2005) Functional analysis of pathogenicity proteins of the potato cyst nematode Globodera rostochiensis using RNAi. Mol. Plant Microb. Interact. 18:621–625
Chen J, Zhang D, Yao Q et al (2010) Feeding based RNA interference of a trehalose phosphate synthase gene in the brown plant hopper, Nilaparvata lugens. Insect Mol Biol 19:777–786
Choudhary D, Koulagi R, Rohatagi D et al (2012) Engineering resistance against root-knot nematode, Meloidogyne incognita, by host delivered RNAi. In: Abstracts of international conference on plant biotechnology for food security: new frontiers. National Agricultural Science Centre, New Delhi, pp 21–24
Christensen J, Litherland K, Faller T et al (2013) Metabolism studies of unformulated internally [3H]- labeled short interfering RNAs in mice. Drug Metab Dispos 41:1211–1219. https://doi.org/10.1124/dmd.112.050666
Christiaens O, Sweveres L, Smagghe G (2014) DsRNA degradation in the pea aphid(Acyrthosiphon pisum) associated with lack of response in RNAi feeding and injection assay. Peptides 53:307–314
Cogoni C, Macino G (2000) Post-transcriptional gene silencing across kingdoms. Curr Opin Genet Dev 10:638–643
Coleman AD, Wouters RH, Mugford ST et al (2015) Persistence and transgenerational effect of plant-mediated RNAi in aphids. JExp Bot 66:541–548
Coy MR, Sanscrainte ND, Chalaire KC et al (2012) Gene silencing in adult Aedes aegypti mosquitoes through oral delivery of double-stranded RNA. J Appl Entomol 136:741–748
Czauderna F, Fechtner M, Dames S et al (2003) Structural variations and stabilizing modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res 31:2705–2716. https://doi.org/10.1093/nar/gkg393
Dalzell JJ, McVeigh P, Warnock et al (2011) RNAi effector diversity in nematodes. PLoS Negl Trop Dis 5:e1176
Danchin EGJ, Arguel MJ, Campan-Fournier A et al (2013) Identification of novel target genes for safer and more specific control of root-knot nematodes from a pan-genome mining. PLoS Pathog 9:e1003745. https://doi.org/10.1371/journal.ppat.1003745
Dang Y, Yang Q, Xue Z et al (2011) RNA interference in fungi: pathways, functions, and applications. Eukaryot Cell 10(9):1148–1155. https://doi.org/10.1128/EC.05109-11
Daniel RGP, John AG (2008) RNAi-mediated crop protection against insects. Trends Biotechnol 26:393–400
Darrington M, Dalmay T, Morrison NI et al (2017) Implementing the sterile insect technique with RNA interference – a review. Entomol Exp Appl 164:155–175. https://doi.org/10.1111/eea.12575
Deng F, Zhao Z (2014) Influence of catalase gene silencing on the survivability of Sitobion avenae. Arch Insect Biochem Physiol 86:46–57
Depicker A, Montagu MV (1997) Post-transcriptional gene silencing in plants. Curr Opin Cell Biol 9:373–382
Dernburg AF, Karpen GH (2002) A chromosome RNAissance. Cell 111:159–162
Dietzl G, Chen D, Schnorrer F et al (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448:151–156
Dinh PTY, Brown CR, Elling AA (2014a) RNA interference of effector gene Mc16D10L confers resistance against Meloidogyne chitwoodi in Arabidopsis and Potato. Phytopathology 104:1098–1106. https://doi.org/10.1094/PHYTO-03-14-0063-R
Dinh PTY, Zhang L, Brown CR et al (2014b) Plant mediated RNA interference of effector gene Mc16D10L confers resistance against Meloidogyne chitwoodi in diverse genetic backgrounds of potato and reduces pathogenicity of nematode offspring. Nematology 6:669–682. https://doi.org/10.1163/15685411-00002796
Dorsett Y, Tuschl T (2004) siRNAs: applications in functional genomics and potential as therapeutics. Nat Rev Drug Discov 3:318–329. https://doi.org/10.1038/nrd1345
Dowling D, Pauli T, Donath A et al (2016) Phylogenetic origin and diversification of RNAi pathway genes in insects. Genome Biol Evol 8:3784–3793. https://doi.org/10.1093/gbe/evw281
Dutta TK, Papolu PK, Banakar P et al (2015) Tomato transgenic plants expressing hairpin construct of a nematode protease gene conferred enhanced resistance to root-knot nematodes. Front Microbiol 6:260. https://doi.org/10.3389/fmicb.2015.00260
Dykxhoorn DM, Novina CD, Sharp PA (2003) Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol 4:457–467
Elbashir SM, Lendeckel W, Tuschl T (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15:188–200
Fairbairn DJ, Cavallaro AS, Bernard M et al (2007) Host-delivered RNAi: an effective strategy to silence genes in plant parasitic nematodes. Planta 226:1525–1533. https://doi.org/10.1007/s00425-007-0588-x
Fan W, Wei Z, Zhang M et al (2015a) Resistance to Ditylenchus destructor infection in sweet potato by the expression of small interfering RNAs targeting unc-15, a movement-related gene. Phytopahol 105:1458–1465. https://doi.org/10.1094/PHYTO-04-15-0087-R
Fan J, Zhang Y, Francis F et al (2015b) Orco mediates olfactory behaviors and winged morph differentiation induced by alarm pheromone in the grain aphid, Sitobion avenae. Insect Biochem Mol Biol 64:16–24
Fanelli E, Di Vito M, Jones JT et al (2005) Analysis of chitin synthase function in a plant parasitic nematode, Meloidogyne artiellia, using RNAi. Gene 349:87–95
Feinberg EH, Hunter CP (2003) Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301:1545–1547
Figueira-Mansur J, Ferreira-Pereira A, Mansur JF et al (2013) Silencing of P-glycoprotein increases mortality in temephos-treated Aedes aegypti larvae. Insect Biochem Mol Biol 22:648–658
Fire A, Xu S, Montgomery MK et al (1998) Potent and specific genetic interference by double-stranded RNA in C. elegans. Nature 391:806–811
Garbian Y, Maori E, Kalev H et al (2012) Bidirectional transfer of RNAi between honey bee and Varroa destructor: Varroa gene silencing reduces Varroa population. PLoS Pathog 8(12):e1003035
Gong L, Yang X, Zhang B et al (2011) Silencing of Rieske iron-sulfur protein using chemically synthesized siRNA as a potential biopesticide against Plutella xylostella. Pest Manag Sci 67:514–520
Gong L, Chen Y, Hu Z et al (2013) Testing insecticidal activity of novel chemically synthesized sirna against Plutella xylostella under laboratory and field conditions. PLoSOne 8:e62990
Gong YH, Yu XR, Shang QL et al (2014) Oral delivery mediated RNA interference of a carboxylesterase gene results in reduced resistance to organophosphorus insecticides in the cotton aphid, Aphis gossypii glover. PLoS One 9:e102823
Good L, Stach JEM (2011) Synthetic RNA silencing in bacteria antimicrobial discovery and resistance breaking. Front Microbiol 2:185
Griebler M, Westerlund SA, Hoffmann KH et al (2008) RNA interference with the allato regulating neuropeptide genes from the fall armyworm Spodoptera frugiperda and its effects on the JH titer in the hemolymph. J Insect Physiol 54:997–1007
Guo H, Song X, Wang G et al (2014) Plant-generated artificial small RNAs mediated aphid resistance. PLoS One 9:e97410
Haegeman A, Joseph S, Gheysen G et al (2011) Analysis of the transcriptome of the root lesion nematode Pratylenchus coffeae generated by 454 sequencing technology. Mol Biochem Parasitol 178:7–14
Hamann L, Jensen K, Harbers K (1993) Consecutive inactivation of both alleles of the gb110 gene has no effect on the proliferation and differentiation of mouse embryonic stem cells. Gene 126:279–284
Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950–952
Hammond SM, Caudy AA, Hannon GJ (2001) Post-transcriptional gene silencing by double-stranded RNA. Nat Rev Genet 2:110–119
He B, Chu Y, Yin M et al (2013) Fluorescent nanoparticle delivered dsRNA toward genetic control of insect pests. Adv Mater Weinheim 25:4580–4584. https://doi.org/10.1002/adma.201301201
Herrero-Vanrell R, Rincón AC, Alonso M et al (2005) Self-assembled particles of an elastin-like polymer as vehicles for controlled drug release. J Control Release 102:113–122. https://doi.org/10.1016/j.jconrel.2004.10.001
Hernandez I, Chacon O, Rodriguez R et al. (2009) Black shank resistant tobacco by silencing of glutathione S- transferase. Biochem Biophys Res Commun 387:300–304
Huang G, Allen R, Davis EL et al (2006) Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc Natl Acad Sci U S A 103:14302–14306. https://doi.org/10.1073/pnas.0604698103
Hull D, Timmons L (2004) Methods for delivery of double-stranded RNA into Caenorhabditis elegans. Methods Mol Biol 265:23–58
Huvenne H, Smagghe G (2010) Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol 56:227–235. https://doi.org/10.1016/j.jinsphys.2009.10.004
Ischizuka A, Siomi MC, Siomi H (2002) A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev 16:2497–2508
Jackson AL, Bartz SR, Schelter J et al (2003) Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 21:635–637. https://doi.org/10.1038/nbt831
Joga MR, Zotti MJ, Smagghe G et al (2016) RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: what we know so far. Front Physiol 7:553. https://doi.org/10.3389/fphys.2016.00553
Kamath RS, Martinez-Campos M, Zipperlen P et al (2001) Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2:2.1–2.10. https://doi.org/10.1186/gb-2000-2-1-research0002
Kennerdell JR, Carthew RW (1998) Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2Act in the wingless pathway. Cell 95:1017–1026
Ketting RF, Fischer SE, Bernstein E et al (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15:2654–2659
Ketting RF (2011) The many faces of RNAi. Dev Cell 15:148-161. https://doi.org/10.1016/j.devcel.2011.01.012
Khajuria C, Buschman LL, Chen MS et al (2010) A gut-specific chitinase gene essential for regulation of chitin content of peritrophic matrix and growth of Ostrinia nubilalis larvae. Insect Biochem Mol Biol 40:621–629. https://doi.org/10.1016/j.ibmb.2010.06.003
Klink VP, Kim KH, Martins V et al (2009) A correlation between host-mediated expression of parasite genes as tandem inverted repeats and abrogation of development of female Heterodera glycines cyst formation during infection of Glycine max. Planta 230:53–71. https://doi.org/10.1007/s00425-009-0926-2
Koch A, Kogel KH (2014) New wind in the sails: improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol J 12:821–831
Koch A, Kumar N, Weber L et al (2013) Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase-encoding genes confers strong resistance to Fusarium species. Proc Natl Acad Sci U S A 110(48):19324–19329. pmid:24218613
Kontogiannatos D, Swevers L, Maenaka K et al (2013) Functional characterization of a juvenile hormone esterase related gene in the moth Sesamia nonagrioides through RNA interference. PLoS One 8:e73834
Kumar M, Gupta GP, Rajam MV (2009) Silencing of acetyl cholinesterase gene of Helicoverpa armigera by siRNA affects larval growth and its life cycle. J Insect Physiol 55:273–278. https://doi.org/10.1016/j.jinsphys.2008.12.005
Kumar A, Wang S, Ou R et al (2013) Development of an RNAi based microalgal larvicide to control mosquitoes. Malaria World J 4:6
Kumar A, Kakrana A, Sirohi A et al (2017) Host-delivered RNAi-mediated root-knot nematode resistance in Arabidopsis by targeting splicing factor and integrase genes. J Gen Plant Pathol 83:91–97. https://doi.org/10.1007/s10327-017-0701-3
Kurreck J (2003) Antisense technologies: improvement through novel chemical modifications. Eur J Biochem 270:1628–1644. https://doi.org/10.1046/j.1432-1033.2003.03555.x
Kwon DH, Park JH, Lee SH (2013) Screening of lethal genes for feeding RNAi by leaf disc-mediated systematic delivery of dsRNA in Tetranychus urticae. Pestic Biochem Physiol 105:69–75. https://doi.org/10.1016/j.pestbp.2012.12.001
Lacroix H, Spanu PD (2009) Silencing of six hydrophobins in Cladosporium fulvum: complexities of simultaneously targeting multiple genes. Appl Environ Microbiol 75:542–546
Lendner M, Doligalska M, Lucius R et al. (2008) Attempts to establish RNA interference in the parasitic nematode Heligmosomoides polygyrus. Mol Biochem Parasitol 161:21–31
Li J, Todd TC, Oakley TR et al (2010a) Host-derived suppression of nematode reproductive and fitness genes decreases fecundity of Heterodera glycines Ichinohe. Planta 232:775–785. https://doi.org/10.1007/s00425-010-1209-7
Li J, Todd TC, Trick HN (2010b) Rapid in planta evaluation of root expressed transgenes in chimeric soybean plants. Plant Cell Rep 29:113–123. https://doi.org/10.1007/s00299-009-0803-2
Li J, Chen Q, Lin Y et al (2011a) RNA interference in Nilaparvata lugens (Homoptera, Delphacidae) based on dsRNA ingestion. Pest Manag Sci 67:852–859
Li X, Zhang M, Zhang H (2011b) RNA interference of four genes in adult Bactrocera dorsalis by feeding their dsRNAs. PLoS One 6:e17788
Li J, Todd TC, Lee J et al (2011c) Biotechnological application of functional genomics towards plant parasitic nematode control. Plant Biotech J 9:936–944. https://doi.org/10.1111/j.1467–7652.2011.00601.x
Li J, Wang XP, Wang MQ et al (2013) Advances in the use of the RNA interference technique in Hemiptera. Insect Sci 20:31–39. https://doi.org/10.1111/j.1744-7917.2012.01550.x
Li XQ, Wei JZ, Tan A et al. (2007) Resistance to root-knot nematode in tomato roots expressing a nematicidal Bacillus thuringiensis crystal protein. Plant Biotechnol J 5:455-464. https://doi.org/10.1111/j.1467-7652.2007.00257.x
Li Y, Wang K, Xie H et al (2015) Cathepsin B cysteine proteinase is essential for the development and pathogenesis of the plant parasitic nematode Radopholus similis. Int J Biol Sci 11:1073–1087. https://doi.org/10.7150/ijbs.12065
Lilley CJ, Goodchild SA, Atkinson HJ et al (2005) Cloning and characterisation of a Heterodera glycines aminopeptidase cDNA. Int J Parasitol 35:1577–1585
Lim LP, Glasner ME, Yekta S et al (2003) Vertebrate micro-RNA genes. Science 299:1540
Lindbo JA, Silva-Rosales L, Proebsting WM et al (1993) Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell 5:1749–1759
Lipardi C, Wei Q, Paterrson BM (2001) RNAi as random degradation PCR: siRNA primers convert mRNA into dsRNA that are degraded to generate new siRNAs. Cell 101:297–307
Lorenz C, Hadwiger P, John M et al (2004) Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg Med Chem Lett 14:4975–4977. https://doi.org/10.1016/j.bmcl.2004.07.018
Lourenço-Tessutti IT, Souza JDA, Martins-de-Sa D et al (2015) Knockdown of heat-shock protein 90 and isocitrate lyase gene expression reduced root-knot nematode reproduction. Phytopathology 105:628–637. https://doi.org/10.1094/PHYTO-09-14-0237-R
Lum L, Yao S, Mozer B et al (2003) Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 299:2039–2045
Luo Y, Wang X, Wang X et al (2013) Differential responses of migratory locusts to systemic RNA interference via double-stranded RNA injection and feeding. Insect Mol Biol 22:574–583. https://doi.org/10.1111/imb.12046
Macrae IJ, Zhou K, Li F et al (2006) Structural basis for double-stranded RNA processing by Dicer. Science 311:195–198
Majumdar R, Rajasekaran K, Cary JW (2017) RNA interference (RNAi) as a potential tool for control of mycotoxin contamination in crop plants: concepts and considerations. Front Plant Sci 8:200. https://doi.org/10.3389/fpls.2017.00200
Manoharan M (2003) RNA interference and chemically modified siRNAs. Nucleic Acids Res Suppl 3:115–116. https://doi.org/10.1093/nass/3.1.115
Mao J, Zeng F (2012) Feeding-based RNA interference of a gap gene is lethal to the pea aphid, Acyrthosiphon pisum. PLoS One 7:e48718
Mao J, Zeng F (2014) Plant-mediated RNAi of a gap gene-enhanced tobacco tolerance against the Myzus persicae. Transgenic Res 23:145–152. https://doi.org/10.1007/s11248-013-9739-y
Mao YB, Cai WJ, Wang JW et al (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol 25:1307–1313. https://doi.org/10.1038/nbt1352
Mao YB, Tao XY, Xue XY et al (2011) Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms. Transgenic Res 20:665–673. https://doi.org/10.1007/s11248-010-9450-1
Mao YB, Xue XY, Tao XY et al (2013) Cysteine protease enhances plant-mediated bollworm RNA interference. Plant Mol Biol 83:119–129. https://doi.org/10.1007/s11103-013-0030-7
Matzke M, Matzke AJ, Kooter JM (2001) RNA: guiding gene silencing. Science 293:1080–1083
Maule AG, McVeigh P, Dalzell JJ et al (2011) An eye on RNAi in nematode parasites. Trends Parasitol 27:505–513. https://doi.org/10.1016/j.pt.2011.07.004
McDonald T, Brown D, Keller NP et al (2005) RNA silencing of mycotoxin production in Aspergillus and Fusarium species. Mol Plant-Microbe Interact 18:539–545
McEwan DL, Weisman AS, Hunter CP (2012) Uptake of extracellular double-stranded RNA by SID-2. Mol Cell 47:746–754. https://doi.org/10.1016/j.molcel.2012.07.014
Meer VRK, Choi MY (2013) Formicidae (ant) control using double-stranded RNA constructs. US Patent No. 8,575,328
Meyering-Vos M, Muller A (2007) RNA interference suggests sulfakinins as satiety effectors in the cricket Gryllus bimaculatus. J Insect Physiol 53:840–848
Miller SC, Miyata K, Brown SJ et al (2012) Dissecting systemic RNA interference in the red flour beetle Tribolium castaneum: parameters affecting the efficiency of RNAi. PLoS One 7:e47431. https://doi.org/10.1371/journal.pone.0047431
Mitter N, Worrall EA, Robinson KE et al (2017) Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nature Plants 3:16207
Miyamoto Y, Masunaka A, Tsuge T et al (2008) Functional analysis of a multicopy host-selective ACT-toxin biosynthesis gene in the tangerine pathotype of Alternaria alternata using RNA silencing. Mol Plant-Microbe Interact 21:1591–1599
Mutti NS, Park Y, Reese JC et al (2006) RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum. J Insect Sci 6:38
Naessens E, Dubreuil G, Giordanengo P et al (2015) A secreted MIF cytokine enables aphid feeding and represses plant immune responses. Curr Biol 25:1898–1903
Nakai K, Horton P (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24:34–36
Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into Petunia results in reversible cosuppression of homologous genes in trans. Plant Cell 2:279–289
Niu JH, Jian H, Xu J et al (2012) RNAi silencing of the Meloidogyne incognita Rpn7 gene reduces nematode parasitic success. Euro J Plant Pathol 134:131–144. https://doi.org/10.1007/s10658-012-9971-y
Niu J, Liu P, Liu Q et al (2016) Msp40 effector of root-knot nematode manipulates plant immunity to facilitate parasitism. Sci Rep 6:19443. https://doi.org/10.1038/srep19443
Nowara D, Gay A, Lacomme C et al (2010) HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22:3130–3141. https://doi.org/10.1105/tpc.110.077040
Nunes CC, Dean RA (2012) Host-induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies. Mol Plant Pathol 13:519–529. https://doi.org/10.1111/J.1364-3703.2011.00766.X
Nunes FMF, Simoes ZLP (2009) A non-invasive method for silencing gene transcription in honeybees maintained under natural conditions. Insect Biochem Mol Biol 39:157–160
Nykanen A, Haley B, Zamore PD (2001) ATP requirement and small interfering RNA structure in the RNA interference pathway. Cell 107:309–321
Ober KA, Jockusch EL (2006) The roles of wingless and decapentaplegic in axis and appendage development in the red flour beetle, Tribolium castaneum. Dev Biol 294:391–405
Panwar V, McCallum B, Bakkeren G (2013) Host-induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the Barley stripe mosaic virus. Plant Mol Biol 81:595–608. https://doi.org/10.1007/s11103-013-0022-7
Papolu PK, Gantasala NP, Kamaraju D et al (2013) Utility of host delivered RNAi of two FMRF amide like peptides, flp-14 and flp-18, for the management of root knot nematode, Meloidogyne incognita. PLoS One 8:e80603. https://doi.org/10.1371/journal.pone.0080603
Pasquinelli AE (2002) MicroRNAs: deviants no longer. Trends Genet 18:171–173
Peng T, Pan Y, Yang C et al (2016) Over-expression of CYP6A2 is associated with spirotetramat resistance and cross-resistance in the resistant strain of Aphis gossypii glover. Pestic Biochem Physiol 126:64–69
Pitino M, Hogenhout SA (2013) Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol Plant-Microbe Interact 26:130–139
Pitino M, Coleman AD, Maffei ME et al (2011) Silencing of aphid genes by dsRNA feeding from plants. PLoSONE 6:e25709. https://doi.org/10.1371/journal.pone.0025709
Possamai SJ, Trionnaire GL, Bonhomme J et al (2007) Gene knockdown by RNAi in the pea aphid Acyrthosiphon pisum. BMC Biotechnol 7:63
Price DR, Gatehouse JA (2008) RNAi-mediated crop protection against insects. Trends Biotechnol 26:393–400. https://doi.org/10.1016/j.tibtech.2008.04.004
Rajagopal R, Sivakumar S, Agrawal N et al (2002) Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as Bacillus thuringiensis toxin receptor. J Biol Chem 277:46849–46851
Rangasamy M, Siegfried BD (2012) Validation of RNA interference in western corn rootworm Diabrotica virgifera virgifera LeConte (Coleoptera, Chrysomelidae) adults. Pest Manag Sci 68:587–591. https://doi.org/10.1002/ps.2301
Rebijith KB, Asokan R, Hande HR et al (2016) RNA interference of odorant-binding protein 2 (OBP2) of the cotton aphid, Aphis gossypii (glover), resulted in altered electrophysiological responses. Appl Biochem Biotechnol 178:251–266
Riechen J (2007) Establishment of broad-spectrum resistance against Blumeria graminis f. sp. tritici in Triticum aestivum by RNAi-mediated knock-down of MLO. J Verbrauch Lebensm 2:120. https://doi.org/10.1007/s00003-007-0282-8
Rodriguez-Cabrera L, Trujillo-Bacallao D, Borra’s-Hidalgo O et al (2010) RNAi-mediated knockdown of a Spodoptera frugiperda trypsin-like serine-protease gene reduces susceptibility to a Bacillus thuringiensis Cry1Ca1 protoxin. Environ Microbiol 12:2894–2903
Romano N, Macino G (1992) Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol 6:3343–3353
Rosso MN, Dubrana MP, Cimbolini N et al (2005) Application of RNA interference to root-knot nematode genes encoding esophageal gland proteins. Mol. Plant Microb. Interact. 18:615–620. https://doi.org/10.1094/MPMI-18-0615
Roxström-Lindquist K, Terenius O, Faye I (2004) Parasite-specific immune response in adult Drosophila melanogaster: a genomic study. Scientific Report 5:207–212. https://doi.org/10.1038/sj.embor.7400073
Santos AC, Sena JAL, Santos SC et al (2009) dsRNA induced gene silencing in Moniliophthora perniciosa, the causal agent of witches’ broom disease of cacao. Fungal Genet Biol 46:825–836
Sapountzis P, Duport G, Balmand S et al (2014) New insight into the RNA interference response against cathepsin-L gene in the pea aphid, Acyrthosiphon pisum: molting or gut phenotypes specifically induced by injection or feeding treatments. Insect Biochem Mol Biol 51:20–32
Schmidt A, Palumbo G, Bozzetti MP et al (1999) Genetic and molecular characterization of sting, a gene involved in crystal formation and meiotic drive in the male germ line of Drosophila melanogaster. Genetics 151:749–760
Senthil-Kumar M, Mysore KS (2011) Caveat of RNAi in plants: the off-target effect. In: Kodama H, Komamine A (eds) RNAi and plant gene function analysis. Methods in molecular biology (methods and protocols), vol 744. Humana Press
Shakesby AJ, Wallace IS, Isaacs HV et al (2009) A water-specific aquaporin involved in aphid osmoregulation. Insect Biochem Mol Biol 39:1–10. https://doi.org/10.1016/j.ibmb.2008.08.008
Shivakumara TN, Sonam C, Divya K et al (2017) Host-induced silencing of two pharyngeal gland genes conferred transcriptional alteration of cell wall-modifying enzymes of Meloidogyne incognita vis-à-vis perturbed nematode infectivity in eggplant. Front Plant Sci 8:473. https://doi.org/10.3389/fpls.2017.00473
Sijen T, Fleenor J, Simmer F et al (2001) On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107:465–476
Sindhu A, Maier TR, Mittchum MG et al (2009) Effective and specific in planta RNAi in cyst nematodes: expression interference of four parasitism genes reduces parasitic success. J Exp Bot 1:315–324. https://doi.org/10.1093/jxb/ern289
Singh AD, Wong S, Ryan CP et al (2013) Oral delivery of double-stranded RNA in larvae of the yellow fever mosquito, Aedes aegypti: implications for pest mosquito control. J Insect Sci 13:69
Siomi H, Siomi MC (2009) On the road to reading the RNA-interference code. Nature 457:396–404. https://doi.org/10.1038/nature07754
Steeves RM, Todd TC, Essig JS et al (2006) Transgenic soybeans expressing siRNAs specific to a major sperm protein gene suppress Heterodera glycines reproduction. Func Plant Biol 33:991–999. https://doi.org/10.1071/FP06130
Surakasi VP, Mohamed AAM, Kim Y (2011) RNA interference of β1 integrin subunit impairs development and immune responses of the beet armyworm, Spodoptera exigua. J Insect Physiol 57:1537–1544
Tabara H, Grishok A, Mello CC (1998) RNAi in C. elegans: soaking in the genome sequence. Science 282:430–431. https://doi.org/10.1126/science.282.5388.430
Tabashnik BE (2008) Delaying insect resistance to transgenic crops. PNAS 105:19029–19030. https://doi.org/10.1073/pnas.0810763106
Tabashnik BE, Gassmann AJ, Crowder DW et al (2008) Insect resistance to Bt crops: evidence versus theory. Nat Biotechnol 26:199–202. https://doi.org/10.1038/nbt1382
Tan FL, Yin JQ (2004) RNAi, a new therapeutic strategy against viral infection. Cell Res 14:460–466
Terenius O, Papanicolaou A, Garbutt JS et al (2011) RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol 57:231–245. https://doi.org/10.1016/j.jinsphys.2010.11.006
Thompson JD, Kornbrust DJ, Foy JW et al (2012) Toxicological and pharmacokinetic properties of chemically modified siRNAs targeting p53 RNA following intravenous administration. Nucleic Acid Ther 22:255–264. https://doi.org/10.1089/nat.2012.0371
Tian H, Peng H, Yao Q et al (2009) Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PLoS One 4:e6225
Timmons L, Court DL, Fire A (2001) Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263:103–112. https://doi.org/10.1016/S0378-1119(00)00579-5
Tinoco ML, Dias BB, Astta RCD et al (2010) In vivo trans-specific gene silencing in fungal cells by in planta expression of a double-stranded RNA. BMC Biol 8:1–11
Tomari Y, Zamore PD (2005) MicroRNA biogenesis: drosha can't cut it without a partner. Curr Biol 15:R61–R64
Tomoyasu Y, Miller SC, Tomita S et al (2008) Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol 9:R10. https://doi.org/10.1186/gb-2008-9-1-r10
Turner CT, Davy MW, MacDiarmid RM et al (2006) RNA interference in the light brown apple moth, Epiphyas postvittana Walker induced by double-stranded RNA feeding. Insect Mol Biol 15:383–391
Tuschl T, Zamore PD, Lehmann R et al (1999) Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev 13:3191–3197
Tzin V, Yang X, Jing X et al (2015) RNA interference against gut osmoregulatory genes in phloem-feeding insects. J Insect Physiol 79:105–112
Ulvila J, Parikka M, Kleino A et al (2006) Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J Biol Chem 281:14370–14375
Upadhyay SK, Chandrashekar K, Thakur N et al (2011) RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J Biosci 36:153–161
Urwin PE, Lilley CJ, Atkinson HJ (2002) Ingestion of double-stranded RNA by pre-parasitic juvenile cyst nematodes leads to RNA interference. Mol Plant Microb Interact 15:747–752. https://doi.org/10.1094/MPMI.2002.15.8.747
Urwin PE, Lilley CJ, McPherson MJ et al. (1997) Resistance to both cyst‐ and root‐knot nematodes conferred by transgenic Arabidopsis expressing a modified plant cystatin. Plant J 12:455–461
Valdes VJ, Sampieri A, Sepulveda J et al (2003) With double stranded RNA to prevent in vitro and in vivo viral infections by recombinant baculovirus. J Biol Chem 278:19317–19324
Van Rij RP, Berezikov E (2009) Small RNAs and the control of transposons and viruses in Drosophila. Trends Microbiol 17:163–171
Vauthier C, Dubernet C, Chauvierre C et al (2003) Drug delivery to resistant tumors: the potential of poly (alkyl cyanoacrylate) nanoparticles. J Control Release 93:151–160. https://doi.org/10.1016/j.jconrel.2003.08.005
Vieira P, Akker EDS, Verma R et al (2015) The Pratylenchus penetrans transcriptome as a source for the development of alternative control strategies: mining for putative genes involved in parasitism and evaluation of in planta RNAi. PLoS One 10:e0144674. https://doi.org/10.1371/journal.pone.0144674
Walawage SL, Britton MT, Leslie CA et al (2013) Stacking resistance to crown gall and nematodes in walnut rootstocks. BMC Genomics 14:668. https://doi.org/10.1186/1471-2164-14-668
Walshe DP, Lehane SM, Lehane MJ (2009) Prolonged gene knockdown in the tsetse fly Glossina by feeding double stranded RNA. Insect Mol Biol 18:11–19
Wang P, Granados RR (2001) Molecular structure of the peritrophic membrane (PM): identification of potential PM target sites for insect control. Arch Insect Biochem Physiol 47:110–118. https://doi.org/10.1002/arch.1041
Wang Y, Zhang H, Li H et al (2011) Second-generation sequencing supply an effective way to screen RNAi targets in large scale for potential application in pest insect control. PLoS One 6:e18644. https://doi.org/10.1371/journal.pone.0018644
Wang W, Luo L, Lu H et al (2015) Angiotensin-converting enzymes modulate aphid–plant interactions. Sci Reports 5:8885
Whyard S, Singh AD, Wong S (2009) Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem Mol Biol 39:824–832. https://doi.org/10.1016/j.ibmb.2009.09.007
Will T, Vilcinskas A (2015) The structural sheath protein of aphids is required for phloem feeding. Insect Biochem Mol Biol 57:34–40
Winston WM, Molodowitch C, Hunter CP et al (2002) Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295:2456–2459
Winston WM, Sutherlin M, Wright AJ et al (2007) Caenorhabditis elegans SID-2 is required for environmental RNA interference. PNAS 104:10565–10570. https://doi.org/10.1073/pnas.0611282104
Winter J, Jung S, Keller S et al (2009) Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11:228–234
Wuriyanghan H, Rosa C, Falk BW (2011) Oral delivery of double-stranded RNAs and siRNAs induces RNAi effects in the potato/tomato psyllid, Bactericera cockerelli. PLoS One 6:e27736
Wynant N, Verlinden H, Breugelmans B et al (2012) Tissue-dependence and sensitivity of the systemic RNA interference response in the desert locust, Schistocerca gregaria. Insect Biochem Mol Biol 42:911–971
Xiao D, Lu YH, Shang QL et al (2015) Gene silencing of two acetylcholinesterases reveals their cholinergic and non-cholinergic functions in Rhopalosiphum padi and Sitobion avenae. Pest Manag Sci 71:523–530
Xiong Y, Zeng H, Zhang Y et al (2013) Silencing the HaHR3 gene by transgenic plant-mediated RNAi to disrupt Helicoverpa armigera development. Int J Biol Sci 9:370–381
Xu HJ, Chen T, Ma XF et al (2013) Genome-wide screening for components of small interfering RNA (siRNA) and micro-RNA (miRNA) pathways in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Mol Biol 22:635–647. https://doi.org/10.1111/imb.12051
Xu L, Duan X, Lv Y et al (2014) Silencing of an aphid carboxylesterase gene by use of plant-mediated RNAi impairs Sitobion avenae tolerance of Phoxim insecticides. Transgenic Res 23:389–396
Xue B, Hamamouch N, Li C et al (2013) The 8D05 parasitism gene of Meloidogyne incognita is required for successful infection of host roots. Phytopathology 103:175–181. https://doi.org/10.1094/PHYTO-07-12-0173-R
Yadav BC, Veluthambi K, Subramaniam K (2006) Host generated double stranded RNA induces RNAi in plant parasitic nematodes and protects the host from infection. Mol Biochem Parasitol 148:219–222. https://doi.org/10.1016/j.molbiopara.2006.03.013
Yanagihara K, Tashiro M, Fukuda Y et al (2006) Effects of short interfering RNA against methicillin-resistant Staphylococcus aureus coagulase in vitro and in vivo. J Antimicrob Chemother 57:122–126
Yang J, Han Z (2014) Efficiency of different methods for dsRNA delivery in cotton bollworm (Helicoverpa armigera). J Integr Agric 13:115–123
Yao J, Rotenberg D, Afsharifar A et al (2013) Development of RNAi methods for Peregrinus maidis, the corn planthopper. PLoS One 8:e370243
Yin C, Jurgenson JE, Hulbert SH (2011) Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol Plant-Microbe Interact 24:554–561. https://doi.org/10.1094/MPMI-10-10-0229
Zamore PD, Tuschl T, Sharp PA (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21- to 23-nucleotide intervals. Cell 101:25–33
Zha W, Peng X, Chen R et al (2011) Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PLoS One 6:e20504. https://doi.org/10.1371/journal.pone.0020504
Zhang Y, Lu Z (2015) Peroxiredoxin 1 protects the pea aphid Acyrthosiphon pisum from oxidative stress induced by Micrococcus luteus infection. J Invertebr Pathol 127:115–121
Zhang H, Kolb F, Jaskiewicz L et al (2004) Single processing center models for human Dicer and bacterial RNase III. Cell 118:57–68
Zhang X, Zhang J, Zhu KY (2010) Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol Biol 19:683–693. https://doi.org/10.1111/j.1365-2583.2010.01029.x
Zhang M, Wang Q, Xu K et al (2011) Production of dsRNA sequences in the host plant is not sufficient to initiate gene silencing in the colonizing oomycete pathogen Phytophthora parasitica. PLoS One 6:e28114. https://doi.org/10.1371/journal.pone.0028114
Zhang Y, Zhang SZ, Kulye M et al (2012) Silencing of molt-regulating transcription factor gene, CiHR3, affects growth and development of sugarcane stem borer, Chilo infuscatellus. J Insect Sci 12:1–12
Zhang H, Li HC, Miao XX (2013a) Feasibility, limitation and possible solutions of RNAi-based technology for insect pest control. Insect Sci 20:15–30
Zhang X, Liu X, Ma J et al (2013b) Silencing of cytochrome P450 CYP6B6 gene of cotton bollworm (Helicoverpa armigera) by RNAi. Bull Entomol Res 103:584–591
Zhang J, Khan SA, Hasse C et al (2015a) Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science 347:991–994
Zhang X, Mysore K, Flannery E et al (2015b) Chitosan/interfering RNA nanoparticle mediated gene silencing in disease vector mosquito larvae. J Vis Exp 97:52523. https://doi.org/10.3791/52523
Zhang J, Khan SA, Heckel DG et al (2017) Next-generation insect-resistant plants: RNAi-mediated crop protection. Trends Biotechnol 35:871–882
Zhao L, Chen J (2013) Double stranded RNA constructs to control ants. US Patent Application Publication No. 2013/0078212
Zhao Y, Yang G, Wang-Pruski G et al (2008) Phyllotreta striolata (Coleoptera, Chrysomelidae): arginine kinase cloning and RNAi-based pest control. Eur J Biochem 105:815–822
Zhou X, Wheeler MM, Oi FM et al (2008) RNA interference in the termite Reticulitermes flavipes through ingestion of double-stranded RNA. Insect Biochem Mol Biol 38:805–815
Zhu F, Xu J, Palli R et al (2011) Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag Sci 67:175–182. https://doi.org/10.1002/ps.2048
Zhu JQ, Liu S, Ma Y et al (2012) Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect-associated gene EcR. PLoS One 7:e38572
Zhuo K, Chen J, Lin B et al (2017) A novel Meloidogyne enterolobii effector MeTCTP promotes parasitism by suppressing programmed cell death in host plants. Mol Plant Pathol 18:45–54. https://doi.org/10.1111/mpp.12374
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Jain, P.K., Bhattacharya, R., Kohli, D., Aminedi, R., Agrawal, P.K. (2018). RNAi for Resistance Against Biotic Stresses in Crop Plants. In: Gosal, S., Wani, S. (eds) Biotechnologies of Crop Improvement, Volume 2. Springer, Cham. https://doi.org/10.1007/978-3-319-90650-8_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-90650-8_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-90649-2
Online ISBN: 978-3-319-90650-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)