Abstract
Plant virus-induced diseases cause significant losses to agricultural crop production worldwide. Reverse genetics systems of plant viruses allow gene manipulation on viral genomes, which greatly facilitates studies of viral pathogenesis and interactions with host organisms. In addition, viral infectious cDNA clones have been modified as versatile recombinant vectors for virus-mediated protein overexpression, virus-induced gene silencing, and gene editing. Since genome RNAs of plant positive-strand RNA viruses are directly translatable, recovery of these viruses has been achieved more than three decades ago by simply expressing viral genome RNA or viral genome-derived in vitro synthesized transcripts in planta. In contrast, genomes of plant negative-strand RNA (NSR) viruses are complementary to their mRNAs and cannot be translated directly. Therefore, rescue of infectious plant NSR viruses from cDNA clones strictly requires the core replication proteins together with their genome RNAs which can assemble into nucleocapsid (NC) complexes as minimal infectious units. However, it is a major challenge to deliver multiple essential components in single cells and to assemble the NC complexes in vivo. Major breakthroughs in reverse genetics systems of plant non-segmented and segmented NSR viruses were just achieved in recent 5 years through various strategies, such as agroinfiltration, minireplicon systems, insect transmission and airbrush inoculation assays. In this review, we summarized critical steps toward developing reverse genetics systems for recovery of several plant NSR viruses in plants and insects. We also highlighted important applications of these reverse genetics of NSR viruses in viral gene function analyses, investigation of virus-insect-plant interactions, and genomic studies of insect vectors and host plants.
Similar content being viewed by others
Background
Plant viruses cause severe crop diseases worldwide and significantly affect annual yield (Scholthof et al. 2011; Jones and Naidu 2019). In the past few decades, plant virologists have developed a growing number of reverse genetics systems of plant viruses for determining viral determinants required for virus replication, movement, host range, and pathogenicity (Jackson and Li 2016; Cody and Scholthof 2019). Furthermore, reverse genetics systems have facilitated studies on virus-host plants, virus-insect vector, and virus-virus interactions. Since plant positive-strand RNA virus genomes are messenger RNAs (mRNAs), reverse genetics systems of these viruses can be achieved by simply introducing in vitro synthesized RNA transcripts into susceptible plant cells. Brome mosaic virus (BMV) was the first plant virus to be rescued from cloned viral cDNA in 1984 (Ahlquist et al. 1984). Since then, reverse genetics systems of numerous plant positive-strand RNA viruses have been developed and exploited as vectors for gene expression and virus-induced RNA silencing (Hefferon 2014; Cody and Scholthof 2019). Thus, significant advances in understanding how plant positive-strand RNA viruses interact with their hosts have been achieved in recent few years.
Negative-stranded RNA (NSR) viruses cause serious diseases in a broad range of hosts, including human, vertebrate, invertebrate, and plant organisms. Plant NSR viruses consist of members of the families Rhabdoviridae, Tospoviridae, Phenuiviridae, Aspiviridae, and Fimoviridae and the unclassified genus Coguvirus (Abudurexiti et al. 2019). Most of plant NSR viruses replicate in arthropod vectors, which facilitates virus transmission and contributes to significant crop yield losses (Jackson et al. 2005; Ammar et al. 2009; Hogenhout et al. 2008; Kormelink et al. 2011). The NSR virus genomes are complementary to their mRNA and have no infectious activity alone. Instead, the minimal infectious units of NSR viruses are nucleocapsids (NCs) or ribonucleoprotein (RNP) complexes, in which the genome RNAs are appropriately associated with the viral RNA polymerase complexes (Jackson et al. 2005; Jackson and Li 2016). Due to difficulties in assembling infectious NCs in vitro from multiple components, development of reverse genetics of NSR virus was lagged behind (Jackson and Li 2016).
Recombinant full-length cDNA clones of a plant nucleorhabdovirus, sonchus yellow net nucleorhabdovirus (SYNV) was rescued successfully in Nicotiana benthamiana plants, which was the first reverse genetics system developed for plant NSR viruses (Ganesan et al. 2013; Wang et al. 2015). Recently, our group have constructed the infectious cDNA clones of a plant cytorhabdovirus, barley yellow striate mosaic virus (BYSMV), which can be rescued in monocot plants and insect vectors (Fang et al. 2019; Gao et al. 2019). Moreover, reverse genetics systems of two plant segmented NSR viruses, tomato spotted wild virus (TSWV) and rose rosette virus (RRV), were established more recently (Pang et al. 2019; Feng et al. 2020). Given rapid research progresses in reverse genetics systems of plant NSR viruses, this review broadly introduces the strategies that have been used for recovery of NSR viruses from cDNA clones and highlights applications of these reverse genetics systems.
Agroinfiltration and minireplicons expressing viral replication proteins and minigenomes: key steps in developing the first plant NSR virus infectious clone
Classical plant rhabdoviruses are non-segmented NSR viruses and consist of the Cytorhabdovirus and Nucleorhabdovirus genera according to their replication and morphogenesis sites on cytoplasm or nucleus. Rhabdovirus genomes encode five structural proteins, including the nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G) and the large protein of viral polymerase (L) (Jackson et al. 2005; Dietzgen et al. 2017). In addition, plant rhabdoviruses encode various accessory genes interspersed the five conserved genes (Walker et al. 2011). The rhabdovirus NC complexes, the minimal infectious units, are composed of the genomic RNA (gRNA) and the viral N protein, the P protein and the L protein. The reverse genetics systems of rhabdoviruses require co-expression of antigenomic RNA (agRNA) with the N, P, and L proteins in single cells and assembly of these components into NC complexes for initiation of virus transcription and replication.
Pioneering works in the development of reverse genetics of NSR viruses were first carried on animal rhabdoviruses. Through co-expression of agRNA with the N, P, and L proteins in trans, Conzelmann and colleagues successfully developed full-length infectious clones of a non-segmented NSR virus, rabies virus (RABV) in 1994 (Schnell et al. 1994). One year later, the recombinant infectious clones of vesicular stomatitis virus (VSV) were constructed (Pattnaik et al. 1995). These systems are excellent models on which we base to construct reverse genetics systems for other NSR viruses infecting animals and plants. However, since plant cell wall interferes with delivery of multiple plasmids into single cells, it is difficult to rescue plant NSR viruses using the same strategies as those used for animal rhabdoviruses (Walpita and Flick 2005; Jackson and Li 2016; German et al. 2020). Furthermore, although NSR viruses can replicate in insect vectors, appropriate insect cell lines are lacking for recovery of plant NSR viruses from their respective infectious clones (Hogenhout et al. 2008).
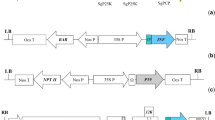
The first reverse genetics system used for recovery of a plant NSR virus was based on SYNV, a nucleorhabdovirus (Ganesan et al. 2013; Wang et al. 2015). In these studies, agroinfiltration and minirepicons (MRs) assays were two key strategies used toward establishing the SYNV infectious cDNA clones (Fig. 1a, b). Agrobacterium tumefaciens-mediated infiltration can efficiently deliver T-DNA into plant cells. Thus, agroinfiltration assays have been utilized for simultaneous co-expression of core replication proteins and genomic RNAs for recovery of plant NSR viruses (Fig. 1a) (Ganesan et al. 2013; Wang et al. 2015). Construction of SYNV MRs was an important step to validate whether the core replicate proteins are functional in virus transcription and replication (Ganesan et al. 2013). The SYNV MR-derivatives were inserted between a hammerhead ribozyme (HHRz) and the hepatitis delta virus ribozyme (HDVRz) sequence to facilitate production of exact genomic termini. In addition, green fluorescent protein (GFP), red fluorescent protein (RFP), and/or chloramphenicol acetyltransferase (CAT) reporter genes replaced the N and P genes to visualize MR transcription and replication in the cells (Fig. 1a). Furthermore, the N, P, and L core proteins of SYNV were also co-expressed by agroinfiltration with A. tumefaciens harboring binary vectors. To improve accumulation of these core proteins and minireplicon derivatives, suppressors of RNA silencing (VSR) proteins, the barley stripe mosaic virus (BSMV) γb, tomato bushy stunt virus (TBSV) p19, and tobacco etch virus (TEV) HC-Pro, were combined into a binary vector to express together in single cells (Ganesan et al. 2013). These MR systems showed that all SYNV core proteins expressed in trans were functional in assembling with MR derivatives, mediating efficient virus transcription and replication in N. benthamiana cells (Ganesan et al. 2013; Jackson and Li 2016).
Combined strategies used in development of reverse genetics systems of plant NSR viruses. a Agrobacterium tumefaciens-mediated expression of genes in N. benthamiana leaves was utilized for simultaneous delivery of plant NSR viruses core protein genes and genomic RNAs in single cells. b The minireplicon (MR) containing a double cauliflower mosaic virus 35S (2 × 35S) promoter, exact termini of the virus, gene junctions, and reporter genes could replicate in conjunction with the ectopic expression of viral core proteins and VSRs, which could validate whether the core replicate proteins are functional in viral transcription and replication. c Full-length viral antigenomic RNA (agRNA) was cloned into the A. tumefaciens binary vector containing a 2 × 35S promoter and a ribozyme (Rz) for cleavage of the 5′ and 3′ ends of the transcribed agRNAs with exact termini. d Insect-mediated transmission of rBYSMV from N. benthamiana leaves to monocot plants. Grinding the infected N. benthamiana leaves and injecting the extracts into the thoraxes of SBPHs. The rBYSMV could be recovered in the SBPHs. After a period of incubation, the infected SBPHs transmitted recombinant viruses to barley plants. e Codon optimization removed cryptic splicing sites in the cDNA of TSWV L and M RNAs for stable expression of the large RdRp protein and genome RNAs in plants. f Inoculation by an airbrush was effective in delivering agrobacterium cultures containing RRV vectors to A. thaliana, N. benthamiana and roses. In the middle panels, combined technologies were used for developing the reverse genetics systems of SYNV, BYSMV, TSWV, and RRV
The successful SYNV MR experiments provided the opportunity to develop the full-length SYNV infectious cDNA clones for use in N. benthamiana plants. The cDNA of SYNV agRNA was engineered between a cauliflower mosaic virus (CaMV) double 35S promoter (2 × 35S) and HΔRz, giving rise to full-length synthetic SYNV agRNA transcripts with authentic 5′- and 3′- ends. (Fig. 1c). The agRNA, N, P, L, and the three VSRs were co-expressed in N. benthamiana leaves through agroinfiltration. At 3 or 4 weeks post infiltration, approximately 5% of the infiltrated plants exhibited classical SYNV symptoms in the systemically infected leaves (Wang et al. 2015). Subsequently, the N, P, and L protein expression cassettes were inserted into a plasmid, which improved the infection efficiency significantly (Wang et al. 2015). The establishment of SYNV reverse genetics has definitely been a template for development of other plant NSR virus infectious cDNA clones (Jackson and Li 2016).
Utilization of insect vectors for recombinant virus transmission is pivotal for rescue of a plant NSR virus in monocot plants and insect vectors
Barley yellow striate mosaic virus, a plant cytorhabdovirus, infects 26 species of cereal plants and is transmitted by the small brown planthopper (SBPH, Laodelphax striatellus) in a propagative manner (Di et al. 2014; Yan et al. 2015; Cao et al. 2018). The BYSMV genome encodes ten proteins including five structural proteins and five accessory proteins in the order of 3′–N–P–P3–P4/P5–P6–M–G–P9–L–5′ (Yan et al. 2015). We have recently developed BYSMV MR and full-length cDNA clones, from which BYSMV can be recovered when inoculated in monocot plants and the insect vectors (Fang et al. 2019; Gao et al. 2019). Different with SYNV, BYSMV cannot infect N. benthamiana plants systemically. However, we found that the BYSMV MRs could establish transcription and replication in agroinfiltrated cells of N. benthamiana plants (Fang et al. 2019). The BYSMV MR experiments provided strong evidence that N. benthamiana plants could support BYSMV replication and transcription in agroinfiltrated cells, and were suitable for recovery of full-length BYSMV. As expected, the full-length BYSMV harboring the RFP reporter gene was recovered from its cDNA clones in agroinfiltrated cells, but cell-to-cell or systemic movement was defective in nonhost N. benthamiana plants (Gao et al. 2019).
To rescue the recombinant BYSMV (rBYSMV) in its natural cereal hosts, we hypothesized that SBPHs could transmit the recombinant viruses from infected N. benthamiana leaves to barley plants (Fig. 1d). However, we found that SBPHs could not transmit the rBYSMV-RFP directly from N. benthamiana leaves to barley plants through conventional feeding method, probably due to that N. benthamiana is nonhost plant of SBPH. Finally, we injected crude extracts obtained from recombinant BYSMV-RFP-infected N. benthaminia leaves into thoraxes of healthy SBPHs. At 8–10 days post injection (dpi), these SBPHs exhibited RFP fluorescence throughout the bodies, indicating that BYSMV-RFP could successfully replicate and disseminate in SBPHs. Then, these BYSMV-RFP-infected SBPHs were transferred to healthy barley plants. At 15 dpi, 90% of the barley plants showed symptoms of chlorotic specks on leaves and stunted growth similar to those observed in wild-type virus-infected barley plants (Gao et al. 2019). We further modified the BYSMV vectors into versatile expression platforms for co-expression of three foreign proteins in cereal plants and SBPHs. Our studies clearly provide new protocols for developing reverse genetics systems of plant NSR viruses infecting monocot plants and arthropod vectors.
Codon optimization is crucial for recovery of a plant segmented NSR virus
Tomato spotted wilt virus is a segmented NSR virus and belongs to the genus Orthotospovirus in the family Tospoviridae (Kormelink et al. 2011; Walker et al. 2019). TSWV is transmitted by thrips in a propagative manner. It infects more than a thousand species of plants and causes enormous economic losses (Pappu et al. 2009; Oliver and Whitfield 2016). TSWV contains a segmented genome, including three RNAs: a large (L), medium (M), and small (S) (Kormelink et al. 2011). The L segment encodes the viral RNA-dependent RNA polymerase (RdRp) in the negative sense. Whereas, the S and M segment are ambisense. The M segment encodes the precursor to two glycolproteins (Gn and Gc) in the negative sense and a nonstructural protein (NSm) in the positive sense. Meanwhile, the S segment encodes a N protein in the negative sense while a nonstructural protein (NSs) in the positive sense (Kormelink et al. 2011). The minimal infectious unit of TSWV consists of viral RNAs, the N protein, and viral RdRp. Only a previous report in 2017 informed the TSWV S RNA synthesis by its RNA polymerase in yeast (Ishibashi et al. 2017). Since the TSWV RdRp is very large (∼330 kDa), it is difficult to express such a large protein in plants through agroinfiltration. In addition, the TSWV L and M gRNAs contain some cryptic splicing sites, leading to the fact that the transcripts of the L and M gRNAs are targeted by splicing systems in the nucleus (Feng et al. 2020). Finally, it is not easy to deliver all the three genome segments and core proteins in single plant cells using agroinfiltration (Feng et al. 2020). Briefly, these difficulties have severely affected the development of reverse genetics system of TSWV in plants.
Recently, Feng et al. have successfully established infectious TSWV entirely from cDNA clones in plants. They firstly developed a S gRNA-based MR system and found it failed though supplemented with N, RdRp proteins and VSRs because of the very low accumulation of RdRp protein. Since numerous intron splicing sites were predicted to exist in the RdRp gene sequence, they then constructed a codon-optimized viral RdRp gene by removing potential splicing sites and used it to stably express the large RdRp protein (Fig. 1e). Consequently, the codon-optimized RdRp, rather than the wild-type RdRp protein, supported transcription and replication of the TSWV S gRNA-based MR system when co-expressed with N protein and four VSRs. Next, the authors constructed a movement-competent MR system based on the M gRNA which contains the NSm protein responsible for cell-to-cell movement, and confirmed that the M gRNA-based MR could move from cell-to-cell in the presence of optimized RdRp and N. Based on these MR systems, they further generated full-length cDNA clones of TSWV. However, no infectious TSWV was recovered in systemically infected leaves due to some splicing sites in the M RNA segment. So, they adapted the same strategy to optimize the M RNA codon sequence. Finally, the TSWV full-length cDNA infectious clones were successfully recovered through co-expression of codon-optimized L, M RNA and wild-type S RNA (Feng et al. 2020). The establishment of this reverse genetics system provides a powerful tool to study life cycles and pathogenicity of tospoviruses.
A reverse genetics system of an emaravirus was developed in rose plants using an airbrush inoculation strategy
Rose rosette virus (RRV) is a negative-sense RNA virus with a 7-segmented genome RNA (Di Bello et al. 2015). It is a member of the Emaravirus genus that includes nine species of virus with four to eight negative-sense RNAs (Di Bello et al. 2015). Among the RRV seven monocistronic genomes, RNA1–4 encode the RdRp, G protein, nucleocapsid, and movement protein, respectively, and RNA1–4 are sufficient for RRV systemic infections. In contrast, The RNA5, RNA6, and RNA7 are not essential for RRV infections (Di Bello et al. 2015). Since RRV is an arthropod-borne virus that is transmitted by eriophyid mites naturally, conventional mechanical inoculation method for RRV transmission into plants is not effective (Pang et al. 2019).
Recently, Pang et al. developed an effective mechanical method by using an airbrush to deliver agrobacterium cultures (Pang et al. 2019). The antigenomic cDNAs of agRNA1 to agRNA7 were introduced between the CaMV 35S promoter and Rz of the binary plasmid pCB301-HDV for viral transcripts with exact 5′ and 3′ termini in vivo. All constructs were transformed into A. tumefaciens and were combined in equal ratios before inoculating into roses using an airbrush method. At 30 and 40 dpi, 88–100% of rose plants exhibited systemic infection symptoms, which was further confirmed by specific RT-PCR assays (Pang et al. 2019). Visual reporter systems were also established by introducing GFP and iLOV protein genes into the RRV infectious cDNA clones (Pang et al. 2019). All these constructs can support recombinant RRV infections in A. thaliana and N. benthamiana plants by agroinfiltration and in roses by the airbrush inoculation approach (Pang et al. 2019). Thus, these new infectious clones facilitate investigation on the molecular mechanisms of RRV in roses.
Generation of recombinant viruses from infectious cDNA clones for studies of virus-plant-insect vector interactions
In the past decades, virus-plant-insect vector interactions have been extensively studied for plant positive-stranded RNA viruses and DNA viruses, which attributes to the availability of their powerful reverse genetics systems. In contrast, the studies of plant NSR viruses and the interaction with their plant host and insect vectors have been heavily constrained due to lack of infectious cDNA clones. Therefore, these newly established reverse genetics systems of plant NSR viruses would allow dissection of detailed molecular mechanisms of virus-host interactions in the context of virus infection. For instance, Sun et al. used the recombinant SYNV mutants to demonstrate that the cooperative G-M interaction is required for inner nuclear membrane invagination and efficient SYNV budding (Sun et al. 2018). Zhou et al. revealed the specific cell-to-cell movement mechanism of plant rhabdoviruses, which requires specific interactions between cognate virus MP and NCs core proteins (Zhou et al. 2019a). They further showed that the matrix protein of SYNV mediates superinfection exclusion (Zhou et al. 2019b).
As obligate parasites, most of plant viruses employ host cytoskeletons for intracellular movement. We recently demonstrated that the BYSMV P protein contributes to the formation of viroplasm-like bodies through recruiting the N and L proteins (Fang et al. 2019). We further confirmed that the P bodies traffic along the ER/actin network driven by myosin XI-K (Fang et al. 2019). Using MR systems of BYSMV, we found that traffic of the BYSMV P protein-formed bodies is required for efficient RNA synthesis in N. benthamiana plants (Fang et al. 2019). Furthermore, our group found that the BYSMV P protein hijacked Carbon catabolite repression 4 (CCR4) to facilitate virus replication in plants and insects (Zhang et al. 2020). The CCR4 proteins were recruited by P into the N-RNA complexes and induced turnover of cellular mRNAs that nonspecifically bind to the BYSMV N protein (Zhang et al. 2020). Since the CCR4 orthologues were conserved in plant and insect hosts, a particularly striking finding is that the virus uses the same trick in both plants and insect vectors. More recently, Gao et al. found that the conserved Casein kinase 1 (CK1) family members regulated cross-kingdom infections of BYSMV in plants and insect vectors by phosphorylating the serine-rich motif that located in an intrinsically disordered region of BYSMV P (Gao et al. 2020). Intriguingly, the CK1-mediated phosphorylation resulted in transformation of BYSMV P from 42 kDa (P42) to 44 kDa, thereby facilitating the transition from virus replication to transcription (Gao et al. 2020).
The establishment of these plant NSR reverse genetics systems would provide powerful tools to generate mutant viruses that will advance our understanding of the molecular arms race in virus-host interactions.
Powerful protein expression platforms based on plant viruses for functional genomics studies in plants and insect vectors
In addition to applications in virus pathogenicity studies, the infectious cDNA clones of plant viruses have been modified into plant virus-mediated overexpression (VOX) vectors for rapid production of foreign proteins. In contrast to time consuming and labor demanding transformation-based methods, VOX-mediated transient expression is rapid and cost-effective (Hefferon 2014; Cody and Scholthof 2019). The VOX-based vectors have long been used to transiently express heterologous proteins with industrial importance in a large-scale production (Hefferon 2014; Cody and Scholthof 2019). Currently, some of these viral vectors have been popularly used to produce plant-derived biopharmaceuticals, such as monoclonal antibodies, vaccines and other therapeutic proteins (Rybicki 2009; Rybicki 2010; Saxena et al. 2016; Hefferon 2017; Cody and Scholthof 2019). In addition, the VOX-based vectors have been widely used for functional characterization of unknown proteins in plant functional genomics studies (Zaidi and Mansoor 2017).
Cauliflower mosaic virus is the first plant virus to be developed as vectors for expression of bacterial genes in 1984 (Brisson et al. 1984). Two years later, the first plant RNA virus vector was developed based on BMV (French et al. 1986). Since then, there have been many successful and popular plant virus-based expression vectors based on positive-strand RNA viruses and some DNA viruses (Palmer and Gleba 2014; Cody and Scholthof 2019). Tobacco mosaic virus (TMV), a rod-shaped virus, is a well characterized plant virus and engineered to be VOX vectors (Cody and Scholthof 2019) which could produce Influenza M2e epitope and human papillomavirus (HPV) E7 protein in plants (Massa et al. 2007; Noris et al. 2011). Subsequently, other rod-shaped viruses including potato virus X (PVX), sun hemp mosaic virus (SHMV) and papaya mosaic potyxvirus (PapMV) were also used to generate epitopes or vaccine (Denis et al. 2008; Liu and Kearney 2010; Hefferon 2017). Geminiviruses, such as beet curly top virus (BCTV), tobacco yellow dwarf virus (TYDV) and bean yellow dwarf virus (BeYDV), are DNA viruses that have been developed as efficient platforms for foreign protein expression (Regnard et al. 2010; Chung et al. 2011; Dugdale et al. 2013). More recently, beet necrotic yellow vein virus (BNYVV) has been engineered as VOX vectors for expression of four foreign proteins in single cells (Jiang et al. 2019). Most of plant VOX vectors could mediate protein overexpression in dicotyledonous plants. In contrast, the development of VOX vectors in monocotyledons has heavily lagged behind. Only a few viruses have been successfully applied in monocots as expression vectors. For instance, BSMV, wheat streak mosaic virus (WSMV), soilborne wheat mosaic virus (SBWMV), and foxtail mosaic virus (FoMV) have been engineered as VOX vectors to express foreign proteins in wheat, barley, oat, and maize plants, respectively (Choi et al. 2000; Lee et al. 2012; Jarugula et al. 2016; Bouton et al. 2018).
Although positive-strand RNA viruses and some DNA viruses have been extensively developed as VOX vectors in both dicotyledonous and monocotyledonous plants, it’s still difficult to stably express large foreign proteins in whole plants due to genetic instabilities of these recombinant viruses (Table 1) (Gleba et al. 2007; Lindbo 2007; Jackson and Li 2016; Cody and Scholthof 2019; Ibrahim et al. 2019). In contrast, non-segmented NSR viruses, like plant rhabdoviruses, were promising VOX vectors, especially for stable expression of large foreign proteins (Jackson and Li 2016; German et al. 2020). Rhabdoviruses have well-defined transcription units and modular genome structure, as well as polar transcription effects (Jackson et al. 2005; Jackson and Li 2016). These features allow them to express foreign proteins regularly, stably and highly. In our recent studies, BYSMV, a plant cytorhabdovirus, was developed as VOX vectors that can simultaneously express three foreign proteins (GFP, RFP, and CFP) in both monocot plants and insect vectors (Gao et al. 2019). Larger proteins like β-glucuronidase (GUS) together with a reporter gene RFP could also been stably expressed in high level in several monocot plants such as barley, wheat, maize and foxtail millet (Gao et al. 2019). Thus, this vector has the potential for other biotechnological applications such as generation of complex antigens, antimicrobial peptides or large and complex pharmaceutical proteins. In addition, this powerful vector can set up a convenient platform for studying the genomic function of plants and insects. The BYSMV-based expression vector was constructed to express RFP and two genes that respectively function in GA biosynthesis (GA5) and inactivation of GA signaling (GA2ox1), showing that high and dwarf phenotypes were induced on barleys overexpressing GA5 or GA2ox1 gene, respectively (Gao et al. 2019). In addition, BYSMV-mediated overexpression of B2, a suppressor of RNA silencing in insect, could improve the BYSMV infections in SBPHs, which indicates that BYSMV expression vectors can be applied in planthoppers (Gao et al. 2019). Therefore, we can easily and efficiently overexpress, knockdown or knockout certain genes in plants and insects to study genomic functions through manipulations of the rBYSMV vector containing visual markers (GFP and RFP).
Application of virus-induced gene editing (VIGE)
Since genome editing mediated by CRISPR-based system has gained widespread popularity, researchers have set their sights on engineering viral vectors for delivery of genome engineering reagents. Currently, Cas9 nucleases and guide RNAs are mainly delivered by transformation to obtain stable transgenic plants or transiently delivery into protoplasts (Cong et al. 2013; Nekrasov et al. 2013). In contrast with these traditional transformative assays, virus vectors provide promising tools for delivery of these gene editing reagents, which would improve the editing efficiency without T-DNA insertions (Table 1) (Cody and Scholthof 2019).
In the first reports of VIGE, geminiviruses and tobacco rattle virus (TRV)-based vectors were utilized for guide RNA delivery in leaves of Cas9-overexpressing N. benthamiana plants. Then, the targeted genome modification was detected in both the inoculated and systemic leaves, even in the seeds of the editing plants, indicating the success of TRV as a genome editing tool (Ali et al. 2015). Subsequently, the VOX vectors of cabbage leaf curl virus (CaLCuV), pea early browning virus (PEBV), TMV, and BNYVV were used for guide RNAs delivery in Cas9 transgenic plants and induce gene editing of endogenous genes in Cas9 transgenic N. benthamiana and/or Arabidopsis plants (Yin et al. 2015; Cody et al. 2017; Ali et al. 2018; Jiang et al. 2019). Recently, BSMV vector could deliver guide RNAs and then induce mutagenesis in both Cas9 transgenic wheat and maize plants (Hu et al. 2019). Nevertheless, the reported success of genome editing is based on Cas9 transgenic plants, because positive strand RNA viruses-based vectors are incapable of carrying the large Cas9 genes.
As described above, plant rhabdoviruses could stably express multiple and large foreign proteins, which expends the research into expression of both the sgRNA and the Cas9 nuclease in one virus vector. As expected, a BYSMV-based vector could simultaneously express the sgRNA and the Cas9 nuclease, which can achieve gene editing in the gfp gene of the N. benthamiana 16c plants (Gao et al. 2019). In addition, the BYSMV vector could express a RFP reporter protein to tract the BYSMV infections and VIGE sites (Gao et al. 2019). More recently, Ma et al. provided a DNA-free plant genome editing tool based on the SYNV vectors. The recombinant SYNV vector carrying a tgtRNA (tRNA-gRNA-tRNA) and a Cas9 was successfully delivered into plants, which resulted in single or multiplex mutagenesis in systemically infected tissues of tobacco plants (Ma et al. 2020). More than 90% of the plants regenerated from virus-infected tissues contained targeted and inheritable mutations (Ma et al. 2020). These results demonstrated that virus vector can actually deliver genome editing reagents and fulfill genetic manipulation without dependence on transgenic lines overexpressing Cas9. Since lots of plants, especially monocot plants, are difficult to be transformed and regenerated, it remains a big challenge to directly infect seed embryo and get inheritable mutations in offspring plants.
Conclusions
Currently, high-throughput RNA sequencing has been employed to identify a growing number of plant viruses that cause damaging losses in agricultural production. However, our understanding of these viruses, especially plant NSR viruses, are far from enough. Here, we highlight key approaches recently used to establish reverse genetics systems of several plant NSR viruses, aiming to give templates for studies of other NSR viruses. Reverse genetics systems of plant NSR viruses will help to solve the fundamental questions of all aspects of viral cycles in the context of natural plant NSR virus infections. For example, our understanding of functions of respective viral protein is not clear. Moreover, insect vectors play an important role in the life cycles of some viruses and obligate virus transmission. But we now know little about the mechanisms behind the insect-virus interaction as well as insect-plant interaction. The establishment of these reverse genetics systems will undoubtedly pave ways for future studies of both virus-insect and vector-plant interactions, and provide new strategies for controlling virus diseases. In addition, these NSR-based vectors are promising tools for foreign protein expression and genome editing in plants or insects (Table 1). The potential of NSR-based virus vector is still underexploited because of lacking effective experimental methods. Last and importantly, it should be emphasized that biosafety should be carefully considered before implementation of these plant virus vectors in fields.
Availability of data and materials
Not applicable.
Abbreviations
- AgRNA:
-
Antigenomic RNA
- BCTV:
-
Beet curly top virus
- BeYDV:
-
Bean yellow dwarf virus
- BMV:
-
Brome mosaic virus
- BNYVV:
-
Beet necrotic yellow vein virus
- BSMV:
-
Barley stripe mosaic virus
- BYSMV:
-
Barley yellow striate mosaic virus
- CaLCuV:
-
Cabbage leaf curl virus
- CaMV:
-
Cauliflower mosaic virus
- CCR4:
-
Carbon catabolite repression 4
- FoMV:
-
Foxtail mosaic virus
- gRNA:
-
Genomic RNA
- mRNA:
-
Messenger RNA
- NC:
-
Nucleocapsid
- NSR virus:
-
Negative-stranded RNA virus
- PapMV:
-
Papaya mosaic potyxvirus
- PEBV:
-
Pea early browning virus
- PVX:
-
Potato virus X
- RABV:
-
Rabies virus
- RdRp:
-
RNA-dependent RNA polymerase
- RNP:
-
Ribonucleoprotein
- RRV:
-
Rose rosette virus
- SBWMV:
-
Soilborne wheat mosaic virus
- SHMV:
-
Sun hemp mosaic virus
- SYNV:
-
Sonchus yellow net virus
- TBSV:
-
Tomato bushy stunt virus
- TEV:
-
Tobacco etch virus
- TMV:
-
Tobacco mosaic virus
- TRV:
-
Tobacco rattle virus
- TSWV:
-
Tomato spotted wild virus
- TYDV:
-
Tobacco yellow dwarf virus
- VIGE:
-
Virus-induced gene editing
- VIGS:
-
Virus-induced gene silencing
- VOX:
-
Virus-mediated protein overexpression
- VSR:
-
Suppressors of RNA silencing
- VSV:
-
Vesicular stomatitis virus
- WSMV:
-
Wheat streak mosaic virus
References
Abudurexiti A, Adkins S, Alioto D, Alkhovsky SV, Avsic-Zupanc T, Ballinger MJ, et al. Taxonomy of the order Bunyavirales: update 2019. Arch Virol. 2019;164:1949–65.
Ahlquist P, French R, Janda M, Loesch-Fries LS. Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proc Natl Acad Sci U S A. 1984;81:7066–70.
Ali Z, Abul-faraj A, Li L, Ghosh N, Piatek M, Mahjoub A, et al. Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol Plant. 2015;8:1288–91.
Ali Z, Eid A, Ali S, Mahfouz MM. Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis. Virus Res. 2018;244:333–7.
Ammar E-D, Tsai C-W, Whitfield AE, Redinbaugh MG, Hogenhout SA. Cellular and molecular aspects of Rhabdovirus interactions with insect and plant hosts. Annu Rev Entomol. 2009;54:447–68.
Bouton C, King RC, Chen H, Azhakanandam K, Bieri S, Hammond-Kosack KE, et al. Foxtail mosaic virus: a viral vector for protein expression in cereals. Plant Physiol. 2018;177:1352–67.
Brisson N, Paszkowski J, Penswick JR, Gronenborn B, Potrykus I, Hohn T. Expression of a bacterial gene in plants by using a viral vector. Nature. 1984;310:511–4.
Cao Q, Xu W-Y, Gao Q, Jiang Z-H, Liu S-Y, Fang X-D, et al. Transmission characteristics of barley yellow striate mosaic virus in its planthopper vector Laodelphax striatellus. Front Microbiol. 2018;9:1419.
Choi IR, Stenger DC, Morris TJ, French R. A plant virus vector for systemic expression of foreign genes in cereals. Plant J. 2000;23:547–55.
Chung HY, Lee HH, Kim KI, Chung HY, Hwang-Bo J, Park JH, et al. Expression of a recombinant chimeric protein of hepatitis a virus VP1-fc using a replicating vector based on beet curly top virus in tobacco leaves and its immunogenicity in mice. Plant Cell Rep. 2011;30:1513–21.
Cody WB, Scholthof HB. Plant virus vectors 3.0: transitioning into synthetic genomics. Annu Rev Phytopathol. 2019;57:211–30.
Cody WB, Scholthof HB, Mirkov TE. Multiplexed gene editing and protein overexpression using a Tobacco mosaic virus viral vector. Plant Physiol. 2017;175:23–35.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23.
Denis J, Acosta-Ramirez E, Zhao Y, Hamelin M-E, Koukavica I, Baz M, et al. Development of a universal influenza a vaccine based on the M2e peptide fused to the papaya mosaic virus (PapMV) vaccine platform. Vaccine. 2008;26:3395–403.
Di Bello PL, Ho T, Tzanetakis IE. The evolution of emaraviruses is becoming more complex: seven segments identified in the causal agent of rose rosette disease. Virus Res. 2015;210:241–4.
Di DP, Zhang YL, Yan C, Yan T, Zhang AH, Yang F, et al. First report of Barley yellow striate mosaic virus on wheat in China. Plant Dis. 2014;98:1450.
Dietzgen RG, Kondo H, Goodin MM, Kurath G, Vasilakis N. The family Rhabdoviridae: mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins. Virus Res. 2017;227:158–70.
Dugdale B, Mortimer CL, Kato M, James TA, Harding RM, Dale JL. In plant activation: an inducible, hyperexpression platform for recombinant protein production in plants. Plant Cell. 2013;25:2429–43.
Fang XD, Yan T, Gao Q, Cao Q, Gao DM, Xu WY, et al. A cytorhabdovirus phosphoprotein forms mobile inclusions trafficked on the actin/ER network for viral RNA synthesis. J Exp Bot. 2019;70:4049–62.
Feng M, Cheng R, Chen M, Guo R, Li L, Feng Z, et al. Rescue of tomato spotted wilt virus entirely from complementary DNA clones. Proc Natl Acad Sci U S A. 2020;117:1181–90.
French R, Janda M, Ahlquist P. Bacterial gene inserted in an engineered RNA virus: efficient expression in monocotyledonous plant cells. Science. 1986;231:1294–7.
Ganesan U, Bragg JN, Deng M, Marr S, Lee MY, Qian S, et al. Construction of a Sonchus yellow net virus minireplicon: a step toward reverse genetic analysis of plant negative-strand RNA viruses. J Virol. 2013;87:10598–611.
Gao Q, Xu W-Y, Yan T, Fang X-D, Cao Q, Zhang Z-J, et al. Rescue of a plant cytorhabdovirus as versatile expression platforms for planthopper and cereal genomic studies. New Phytol. 2019;223:2120–33.
Gao Q, Yan T, Zhang Z-J, Liu S-Y, Fang X-D, Gao D-M, et al. Casein kinase 1 regulates cytorhabdovirus replication and transcription by phosphorylating a phosphoprotein serine-rich motif. Plant Cell. 2020. https://doi.org/10.1105/tpc.1120.00369.
German TL, Lorenzen MD, Grubbs N, Whitfield AE. New technologies for studying negative-strand RNA viruses in plant and arthropod hosts. Mol Plant-Microbe Interact. 2020;33:382–93.
Gleba Y, Klimyuk V, Marillonnet S. Viral vectors for the expression of proteins in plants. Curr Opin Biotech. 2007;18:134–41.
Hefferon K. Plant virus expression vector development: new perspectives. Biomed Res Int. 2014;2014:785382.
Hefferon K. Plant virus expression vectors: a powerhouse for global health. Biomedicines. 2017;5:44.
Hogenhout SA, Ammar E-D, Whitfield AE, Redinbaugh MG. Insect vector interactions with persistently transmitted viruses. Annu Rev Phytopathol. 2008;46:327–59.
Hu J, Li S, Li Z, Li H, Song W, Zhao H, et al. A barley stripe mosaic virus-based guide RNA delivery system for targeted mutagenesis in wheat and maize. Mol Plant Pathol. 2019;20:1463–74.
Ibrahim A, Odon V, Kormelink R. Plant viruses in plant molecular pharming: toward the use of enveloped viruses. Front Plant Sci. 2019;10:803.
Ishibashi K, Matsumoto-Yokoyama E, Ishikawa M. A tomato spotted wilt virus S RNA-based replicon system in yeast. Sci Rep. 2017;7:12647.
Jackson AO, Dietzgen RG, Goodin MM, Bragg JN, Deng M. Biology of plant rhabdoviruses. Annu Rev Phytopathol. 2005;43:623–60.
Jackson AO, Li Z. Developments in plant negative-strand RNA virus reverse genetics. Annu Rev Phytopathol. 2016;54:469–98.
Jarugula S, Charlesworth SR, Qu F, Stewart LR. Soil-borne wheat mosaic virus infectious clone and manipulation for gene-carrying capacity. Arch Virol. 2016;161:2291–7.
Jiang N, Zhang C, Liu JY, Guo ZH, Zhang ZY, Han CG, et al. Development of Beet necrotic yellow vein virus-based vectors for multiple-gene expression and guide RNA delivery in plant genome editing. Plant Biotechnol J. 2019;17:1302–15.
Jones RAC, Naidu RA. Global dimensions of plant virus diseases: current status and future perspectives. Annu Rev Virol. 2019;6:387–409.
Kormelink R, Garcia ML, Goodin M, Sasaya T, Haenni AL. Negative-strand RNA viruses: the plant-infecting counterparts. Virus Res. 2011;162:184–202.
Lee WS, Hammond-Kosack KE, Kanyuka K. Barley stripe mosaic virus-mediated tools for investigating gene function in cereal plants and their pathogens: virus-induced gene silencing, host-mediated gene silencing, and virus-mediated overexpression of heterologous protein. Plant Physiol. 2012;160:582–90.
Lindbo JA. High-efficiency protein expression in plants from agroinfection-compatible Tobacco mosaic virus expression vectors. BMC Biotechnol. 2007;7:52.
Liu Z, Kearney CM. A tobamovirus expression vector for agroinfection of legumes and Nicotiana. J Biotechnol. 2010;147:151–9.
Ma X, Zhang X, Liu H, Li Z. Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat Plants. 2020;6:773–9.
Massa S, Franconi R, Brandi R, Muller A, Mett V, Yusibov V, et al. Anti-cancer activity of plant-produced HPV16 E7 vaccine. Vaccine. 2007;25:3018–21.
Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:691–3.
Noris E, Poli A, Cojoca R, Rittà M, Cavallo F, Vaglio S, et al. A human papillomavirus 8 E7 protein produced in plants is able to trigger the mouse immune system and delay the development of skin lesions. Arch Virol. 2011;156:587–95.
Oliver JE, Whitfield AE. The genus Tospovirus: emerging Bunyaviruses that threaten food security. Annu Rev Virol. 2016;3:101–24.
Palmer K, Gleba Y. Plant viral vectors. Heidelberg: Springer; 2014.
Pang M, Gayral M, Lyle K, Shires MK, Ong K, Byrne D, et al. Infectious DNA clone technology and inoculation strategy for Rose Rosette Virus that includes all seven segments of the negative-strand RNA genome. bioRxiv. 2019:712000.
Pappu HR, Jones RAC, Jain RK. Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res. 2009;141:219–36.
Pattnaik AK, Ball LA, Legrone A, Wertz GW. The termini of VSV DI particle RNAs are sufficient to signal RNA encapsidation, replication, and budding to generate infectious particles. Virology. 1995;206:760–4.
Regnard GL, Halley-Stott RP, Tanzer FL, Hitzeroth II, Rybicki EP. High level protein expression in plants through the use of a novel autonomously replicating geminivirus shuttle vector. Plant Biotechnol J. 2010;8:38–46.
Rybicki EP. Plant-produced vaccines: promise and reality. Drug Discov Today. 2009;14:16–24.
Rybicki EP. Plant-made vaccines for humans and animals. Plant Biotechnol J. 2010;8:620–37.
Saxena P, Thuenemann EC, Sainsbury F, Lomonossoff GP. Virus-derived vectors for the expression of multiple proteins in plants. In: MacDonald J, Kolotilin I, Menassa R, editors. Recombinant proteins from plants. Methods in molecular biology, vol. 1385. New York: Humana Press; 2016. p. 39–54.
Schnell MJ, Mebatsion T, Conzelmann KK. Infectious rabies viruses from cloned cDNA. EMBO J. 1994;13:4195–203.
Scholthof K-BG, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, et al. Top 10 plant viruses in molecular plant pathology. Mol Plant Pathol. 2011;12:938–54.
Sun K, Zhou X, Lin W, Zhou X, Jackson AO, Li Z. Matrix-glycoprotein interactions required for budding of a plant nucleorhabdovirus and induction of inner nuclear membrane invagination. Mol Plant Pathol. 2018;19:2288–301.
Walker PJ, Dietzgen RG, Joubert DA, Blasdell KR. Rhabdovirus accessory genes. Virus Res. 2011;162:110–25.
Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Dempsey DM, Dutilh BE, et al. Changes to virus taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses. Arch Virol. 2019;164:2417–29.
Walpita P, Flick R. Reverse genetics of negative-stranded RNA viruses: a global perspective. FEMS Microbiol Lett. 2005;244:9–18.
Wang Q, Ma X, Qian S, Zhou X, Sun K, Chen X, et al. Rescue of a plant negative-strand RNA virus from cloned cDNA: insights into enveloped plant virus movement and morphogenesis. PLoS Pathog. 2015;11:e1005223.
Yan T, Zhu JR, Di D, Gao Q, Zhang Y, Zhang A, et al. Characterization of the complete genome of barley yellow striate mosaic virus reveals a nested gene encoding a small hydrophobic protein. Virology. 2015;478:112–22.
Yin K, Han T, Liu G, Chen T, Wang Y, Yu AY, et al. A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci Rep. 2015;5:14926.
Zaidi SS-e-A, Mansoor S. Viral vectors for plant genome engineering. Front Plant Sci. 2017;8:539.
Zhang Z-J, Gao Q, Fang X-D, Ding Z-H, Gao D-M, Xu W-Y, et al. CCR4, a RNA decay factor, is hijacked by a plant cytorhabdovirus phosphoprotein to facilitate virus replication. eLife. 2020;9:e53753.
Zhou X, Lin W, Sun K, Wang S, Zhou X, Jackson AO, et al. Specificity of plant Rhabdovirus cell-to-cell movement. J Virol. 2019a;93:e00296–19.
Zhou X, Sun K, Zhou X, Jackson AO, Li Z. The matrix protein of a plant Rhabdovirus mediates superinfection exclusion by Inhibiting viral transcription. J Virol. 2019b;93:e00680-19.
Acknowledgements
We thank all the members of molecular plant virology (MPV) lab in CAU for their helpful suggestions and constructive criticism.
Funding
This work was supported by the Natural Science Foundation of China (Grants 31872920 and 31571978 to XBW).
Author information
Authors and Affiliations
Contributions
YZ and XBW drafted the manuscript. YZ and XBW designed and made the Figure and Table. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zang, Y., Fang, XD., Qiao, JH. et al. Reverse genetics systems of plant negative-strand RNA viruses are difficult to be developed but powerful for virus-host interaction studies and virus-based vector applications. Phytopathol Res 2, 29 (2020). https://doi.org/10.1186/s42483-020-00068-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42483-020-00068-5