Abstract
Bioherbicides afford satisfactory approach to overcome weed problem. Selection of new bioherbicides from phytopathogenic microbiota is essential for modern agricultural management, especially mycoherbicides. Thus, in the present investigation, 4 phytopathogenic fungi including Alternaria alternata YMM1, A. tenuissima YMM3, Chaetomium globosum YMM2, and Nigrospora oryzae YMM4 were isolated from infected leaves with chlorotic rings of Rumex dentatus and Sonchus oleraceus as common weeds in Egypt. These fungi were identified based on morphology and sequence analysis of ITS1-5.8SrDNA-ITS2 of rRNA genes. Mycoherbicidal metabolites were produced using submerged cultures in potato dextrose broth. Their bioherbicidal activities were evaluated using seed germination and leaf disk puncture assays. It was found that the most promising fungal strain against major tested weeds was N. oryzae YMM4. Thus, phytopathogenic microbiota are considered a great resource for the construction of a new bioherbicide for managing weeds.
Similar content being viewed by others
Background
Weeds are the main problem in intensive agriculture and compete for the same nutrients of crop plants (Sahin, 2019). Therefore, the quality of crops and yields has decreased significantly and caused enormous economic losses (Bajwa et al. 2018). Weed control is vital to increase crops’ yields. Management approaches mainly rely on synthetic herbicide systems for increasing crop productivity (Cordeau et al. 2016). However, synthetic chemicals are inadequate due to their resistance to degradation, environmental risks, and the development of resistant varieties of weeds (Schütte et al. 2017). Resistant individuals among weed populations are developed through the intensive uses of the same synthetic herbicides for long times (Matthews 2018). Hence, these synthetic herbicides need to be replaced with environmentally friendly biological herbicides (bioherbicides).
Bioherbicides are ecofriendly materials and usually have shorter half-life than synthetic herbicides (Javed et al. 2016). Fungi are well investigated to bioherbicidal production for weedy management (Guo et al. 2020). Phytopathogenic molds generate metabolites that play a great role in the improvement of weed control. Production of bioherbicides from plant pathogens has been confirmed to be an effective approach for weed control (Júnior et al. 2019). Broth cultures of several fungi showed considerable herbicidal activity in both crude and purified forms against various weeds (Sica et al. 2016). Bioherbicidal production for management of weeds involves a sequence of steps, including isolation and selection of phytopathogenic fungi from infected weeds and broth medium were used for bioherbicides production (de Souza et al. 2015).
The phytopathogenic fungal biodiversity in diseased weeds of cultivated areas in Egyptian agriculture has been seldom studied, especially for production of bioactive metabolites. Therefore, the main objective of the present study was to isolate phytopathogenic fungi from infected leaves with chlorotic rings of Rumex dentatus and Sonchus oleraceus as common weeds in Egypt. In addition, in vitro cultivation of the isolated phytopathogenic fungi in the laboratory was done to obtain filtrates containing biologically active metabolites with herbicidal action and evaluate their herbicidal effects against some weeds.
Materials and methods
Culture media
Acetonitrile, methanol, and water of HPLC grade were purchased from Sigma-Aldrich Co. (Spruce Street, St. Louis, MO, USA). Glucose, magnesium sulphate, yeast extract, peptone, beef extract, dimethyl sulfoxide (DMSO), and agar were supplied from El-Gomhoria for pharmaceutical and chemicals Co., Alexandria, Egypt. Potato dextrose agar (PDA) was purchased from Oxoid (Basingstoke, Hampshire, RG24 8PW, UK). Other chemicals and solvents were purchased from El-Nasr Pharmaceutical Chemicals Co., Qalyub, Egypt, and used without further purification. All media were prepared immediately and autoclaved at 121 °C for 20 min.
Sampling area and collection of weed specimens
Plant weeds (Rumex dentatus and Sonchus oleraceus) with visible diseased symptoms of samples were collected from wheat field and placed in sterilized glass bottles. The symptoms of all diseased weeds were characterized by necrotic (an irregular brownish) spots with chlorotic halo around them. The seeds and leaves used in the present study were collected from weeds growing in fields of cultivated crops. Sampling locality was a cultivated area at Jarrar, Abu Hommos, Beheira Governorate, Egypt. The weed species were identified by the help of available literature (Täckholm 1974; Boulos 1999, 2002, 2005 and 2009).
Isolation and selection of phytopathogenic fungi
Phytopathogenic fungi were isolated from the infected leaves of R. dentatus and S. oleraceus weeds. The leaves were surface sterilized by 95% ethyl alcohol for 1 min, then dipped in 10% sodium hypochlorite for one minute, followed by washing in sterile water. The infected spots of leaves were directly transferred to a Petri plate containing PDA medium. Inoculated plates were incubated at 28 °C and observed after 7 days (Babu et al. 2003 and Aybeke 2017). Fungal colonies were selected for purification by repeated streaking on agar plates of the PDA. The pure colonies obtained were transferred to a fresh PDA slant, subcultured, and stored at 4 °C.
Identification of phytopathogenic fungi
Phytopathogenic fungi were identified morphologically on the basis of macroscopic (naked eye) and microscopic characteristics (Moubasher 1993) after culturing on the PDA medium at 28 °C for 7 days. For molecular characterizations of fungal isolates, total genomic DNA preparations of fungal cultures in potato dextrose broth (PDB) were made according to the protocol of Quick-DNA™ Fungal Microprep Kit (Zymo research #D6007). The extracted DNA preparations were examined using 1% agarose gel electrophoresis. The internal transcribed spacer (ITS) region (ITS1-5.8SrDNA-ITS2) of rRNA genes was amplified, using the primer pairs of ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) (White et al. 1990). The 5.8S rRNA gene and the two flanking internal transcribed spacer regions (ITS1-5.8S rDNA-ITS2) of rRNA genes were PCR amplified according to the protocol of Maxima Hot Start PCR Master Mix (Thermo K1051). The amplifications were performed in a ThermoHybaid PCR Sprint Thermal Cycler (Thermo Electron, USA). PCR products were purified according to the protocol of GeneJET™ PCR Purification Kit (Thermo K0701). Amplicons, along with the marker DNA (DNA Ladder), were visualized by 1 % agarose gel electrophoresis after staining with ethidium bromide to confirm the size and purity of the amplified PCR products. The amplified PCR products of ITS1-5.8SrDNA-ITS2 were sequenced on both strands using ITS1 and ITS4 primers at GATC Company by use ABI 3730xl DNA sequencer. The nucleotide sequences determined were deposited in GenBank. For the identification of the isolates, the nucleotide sequences obtained were compared with those sequences already deposited in the data bank of the National Centre for Biotechnology and Information (NCBI) using the nucleotide basic local alignment search tool (BLASTn) to find the most closely related sequences. The identification of the species was determined based on the best sequence alignment score. The obtained DNA fragments were also subjected to a phylogenetic study by means of comparative sequence analysis of the ITS regions including the 5.8S rDNA sequences. This was achieved by generating a neighbor-joining distance-based tree using the software MEGA 6.0.
Preparation of fungal inocula
For inoculum preparations, PDA Petri plates were inoculated by phytopathogenic fungi from slants and incubated at 28 °C for 7 days. Then, the culture surfaces on the agar plates were scraped with 10 ml of sterilized water, using isolation needle. The spores were obtained and the suspensions were counted using a hemocytometer to obtain the desired concentrations of 2.5 × 105 conidia/ml and used as inoculum sources.
Preparation of fungal bioactive metabolites with herbicidal action
For the production of fungal filtrates with herbicidal activity, the fungal culture broths were prepared using 250-ml Erlenmeyer conical flasks containing 100 ml of PDB medium inoculated by 2 ml of spore suspension (2.5 × 105 conidia/ml) at pH 6 and in static conditions at 28 °C (Babu et al. 2003 and Guo et al. 2020). After 7 days of incubation, fungal mycelia were separated by filtration on a pre-weighed Whatman filter paper (No. 4), washing twice with distilled water and dried to a constant weight at 60 °C and reweighed. Fungal dry biomass was measured gravimetrically as the difference in weight. The culture filtrate for each fungus (≈ 500 ml) was centrifuged at 10,000 rpm for 10 min and the supernatant was lyophilized.
Bioherbicidal activity of crude fungal filtrates
The lyophilized product (0.1 g) that was obtained from the fungal filtrate was dissolved in 50 ml distilled water and sterilized, using 0.45 μm Minisart membrane filter.
In vitro bioherbicidal activity
The in vitro bioherbicidal activities of the filtrates of phytopathogenic fungi were assayed by seed germination bioassay and seedling morphology. All seeds were treated by 0.5% sodium hypochlorite for 10 min and washed numerous times with sterile distilled water before germination assay immediately.
Evaluation of bioherbicidal potential of crude filtrate, using an in vitro seed germination bioassay was carried out on filter paper (Whatman No. 4) in Petri dishes (10 cm in diameter) against R. dentatus L., S. oleraceus L., Avena fatua L., Polypogon monspeliensis L., Setaria viridis L., Echinochloa crus-galli L. Beauv, E. colona L., and Plantago major L. seeds. Crude filtrate of each phytopathogenic fungus (2 ml daily) was poured on the filter paper in Petri plate containing 100 seeds and incubated for 10 days under alternation between light (12 h using 2000 Lux) and dark (12 h) at 25 °C. Germination (%), plumule heights, and radical lengths of seedlings were examined. Control treatments were carried out, using sterile PDB medium instead of crude filtrates. Each trial was examined with three replications.
In vivo bioherbicidal activity
The in vivo bioherbicidal activities of crude filtrates were tested by leaf disk puncture assay against leaves of the abovementioned weeds. Crude filtrate of each phytopathogenic fungus (20 μL) was applied on leaf disks of the selected plants. The disks (13-mm diameter) were cut from weed leaves, then located on wetted filter paper (Whatman No. 4) within Petri plates, and punctured by sterile needle in the center before addition of fungal filtrates. Leaf disks were kept under constant light conditions and 25 °C. After 5 days of incubation, symptoms were examined visually. The degree of phytotoxicity (%) was scored according to severity of the symptoms on leaf disks. Control treatments were carried out, using sterile PDB medium instead of crude filtrates (Guo et al. 2020). Each trial was examined with three replications.
Isolation of fungal extracts and GC/MS analysis
For preparation of fungal extracts, culture filtrates of phytopathogenic fungi (500 ml) were lyophilized. A weight of 0.1 g was dissolved in 50 ml distilled water and stored at 4 °C in the dark. The compositions of the extracts were analyzed (Ekman & Holmbom 1989) by gas chromatography/mass spectrometry (GC/MS) with the following specifications: A Trace GC Ultra/Mass Spectrophotometer ISQ (Thermo Scientific) instrument was equipped with flame ionization detector (FID) and a DB-5 narrow bore column. Helium (average velocity 39 cm s−1) was used as the carrier gas (flow rate of 1 ml/min), and the temperature program was 120 °C/min, raised at 6 °C/min to 320 °C, injector temperature was 260 °C, and detector temperature was 320 °C with post run (off) at 320 °C. The GC/MS was equipped with a ZB-5MS Zebron capillary column (length 30 m × 0.25 mm ID, 0.25-μm film thickness; Agilent). A sample (1 μl) was injected at 250 °C, with split/split-less injector (50:1 split ratio) in the split-less mode flow with 10 ml/min. All mass spectra were recorded in the electron impact ionization (EI) at 70 electron volts. The mass spectrometer was scanned from 50 to 500 m/z at five scans per second. Scan time is 1.5 s and mass range is 40 to 300 amu. Peak area percent was used for obtaining quantitative data with the Xcalibur software (Thermo Scientific) without the use of response factor correction. Constituents were identified by comparing their mass spectra with MS library (NIST, Mainlib, Reblib and Wiley) data (Adams 1995). Quantification of constituents was obtained by integrating the peak area of the chromatogram.
Statistical analysis
Experimental data are presented as mean ± standard error and the analysis of variance (ANOVA) of data was conducted and mean property values were compared (p ≤ 0.05) to Fisher’s least significant difference (LSD) method by Minitab 16.1.0 program (Minitab Inc., PA, USA).
Results and discussion
Fungal isolation and characterizations
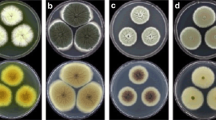
Four phytopathogenic fungal strains were isolated from infected lesions of weed leaves. YMM1 and YMM2 were isolated from R. dentatus and YMM3 and YMM4 from S. oleraceus. These fungal isolates were identified based on morphology and phylogenetic analysis. According to morphological features, the isolates belonged to 3 different genera, Alternaria (YMM1 and YMM3), Chaetomium (YMM2), and Nigrospora (YMM4).
Purified genomic DNA was prepared for PCR to amplify the ITS regions, using ITS1 and ITS4 primers, which resulted in fragments, ranging in length between 531 and 610 bp (Fig. 1). The same primers were then separately used for nucleotide sequencing of the purified DNA fragments. After editing, the obtained sequences were submitted to the GenBank and the received accession numbers were shown in Table 1. ITS regions of rDNA are widely used to study the variability in fungi at the species levels (Bonito et al. 2010 and Mohankumar et al. 2010).
The database matching results of the sequences obtained in the present study confirmed that the 4 examined isolates belonged to 3 different genera (Table 1). The observed high identity results (99 %) to database sequences allowed the identification of the isolates at the species level. These included 2 Alternaria spp. (A. alternata YMM1 and A. tenuissima YMM3), one Chaetomium sp. (C. globosum YMM2), and one Nigrospora sp. (N. oryzae YMM4).
In a similar study, A. alternata was isolated from diseased leaves of Lantana camara (Saxena & Pandey 2002). In addition, a phytopathogenic fungus Septoria polygonorum was isolated from sporulating lesions on Polygonum pensylvanicum weed (Mitchell 2003). Five isolates of Phoma spp. were isolated from Setaria viridis (Zhang et al. 2003). Fusarium oxysporum was isolated from Striga hermonthica stems (Marley & Shebayan 2005). Colletotrichum graminicola and Gloeocercospora sorghi were collected from sporulating lesions on Johnson grass and were used for the control of shattercane weed (Mitchell et al. 2008). Phyllosticta cirsii, a phytopathogen was isolated from diseased leaves of Cirsium arvense (Tuzi et al. 2010).
Based on the ITS of rDNA sequences obtained from the experimental isolates and their close relatives, a phylogenetic neighbor-joining tree that reflected evolutionary relationships was constructed for each of the three genera using MEGA 6.0 (Fig. 2).
Effect of fungal filtrates on germination and growth parameters of weeds
In the present study, weed species competing with 2 major crops (wheat and rice) were selected. The ability of fungal cultures filtrates to inhibit seed germination and seedling growth of different weeds was examined and compared by controls. Data in Table 2 demonstrated that all crude filtrates from the 4 phytopathogenic fungal strains significantly reduced radical lengths and plumule heights of R. dentatus, A. fatua, P. monspeliensis, S. viridis, S. oleraceus, and P. major than the controls. In addition, crude filtrate of N. oryzae YMM4 showed the strongest reduction effect against seedling elevation and seed germination (%) of major tested plants. Furthermore, seedlings of E. colona and E. crus-galli were sensitive and significantly inhibited by crude filtrate of N. oryzae YMM4. In addition, culture filtrate of N. oryzae YMM4 was the most active agent against weeds seeds germination. No seed germination occurred in case of E. crus-galli treated by crude filtrate of N. oryzae YMM4. However, culture filtrates of A. tenuissima YMM3 and C. globosum YMM2 improved seedling growth of E. crus-galli and E. colona, respectively.
Previous studies confirmed the bioherbicidal activities of culture filtrates of diverse phytopathogenic genera including: Drechslera, Diaporthe, Chaetomium, Alternaria, and Nigrospora against different weed species (Saxena & Pandey 2002; Babu et al. 2003; Shabana 2005; Lee et al. 2008; Mitchell et al. 2008; Varejão et al. 2013; Akbar & Javaid 2015; de Souza et al. 2015; de Oliveira Bastos et al. 2017). Zhang et al. (2013) found that the secondary metabolites of the Pythium aphanidermatum including 4-hydroxy-3-methoxycinnamic acid and indole derivatives showed an herbicidal activity on the radical and coleoptile elongation of Digtaria sanguinalis. Akbar and Javaid (2015) concluded that cultural filtrates of Drechslera australiensis and D. hawaiiensis contain herbicidal metabolites that resulted in about 58% reduction in biomass of R. dentatus. Coniolariella sp. produced mevalocidin, a secondary metabolite phytotoxin that revealed a wide range of herbicidal actions (Sica et al. 2016).
Bioherbicidal activity of fungal filtrates on weed leaves
According to leaf disk puncture bioassay; the herbicidal effects of fungal crude filtrates varied according to plant species and filtrate type, and a variety of the symptoms appeared including chlorosis and necrosis. Culture filtrates of A. alternata YMM1 and C. globosum YMM2 caused severe injury symptoms against leaf disks of P. major, P. monspeliensis, and R. dentatus. Also, they cause mild injury symptoms against S. oleraceus and E. crus-galli, respectively. Similarly, culture filtrates of A. tenuissima YMM3 showed sever injury symptoms against leaf disks of P. monspeliensis and mild injury symptoms against R. dentatus and P. major. Likewise, culture filtrate of N. oryzae YMM4 exhibited severe injury symptoms against leaf disks of E. crus-galli and mild injury symptoms against P. major (Table 2). Similar symptoms were observed in several previous studies by bioherbicides (Varejão et al. 2013; de Souza et al. 2015 and Pes et al. 2016). In the present study, culture filtrates of fungal species had a non-selective toxicity against a wide range of tested weeds. These activities may be due to the toxins produced by phytopathogens and cause disease symptoms.
Characterizations of fungal metabolites in crude extracts
GC/MS analyses of the crude extracts from A. alternata YMM1, C. globosum YMM2, A. tenuissima YMM3, and N. oryzae YMM4 (Fig. 3) led to the identification of 15, 22, 23, and 15 different components, representing 70.98, 91.27, 66.46, and 82.87% of the total extracts for the tested fungi, respectively. The identified compounds were tested according to their elution order on a ZB-5MS Zebron capillary column. The results showed some constituents’ similarities among extracts of the 4 tested fungi.
Conclusion
Four fungal phytopathogens A. alternata YMM1, C. globosum YMM2, A. tenuissima YMM3, and N. oryzae YMM4 were isolated from R. dentatus and S. oleraceus weeds and their bioherbicidal activities were characterized. Their broth cultures showed a strong phytotoxic activity on the tested weeds. Within these 4 phytopathogenic fungi, N. oryzae YMM4 showed the greatest bioherbicidal activity. This genus has previously been recorded as a producer of bioherbicidal metabolites. Thus, N. oryzae YMM4 metabolites might be used as a bioherbicide to control several weeds in Egypt. In the future, it would be of interest to further investigate the potential of this isolate under controlled conditions and/or field, such as those underway in Egypt, as a biological control against some weeds to confirm the current results.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author, Youssef M. M. Mohammed, upon reasonable request.
Abbreviations
- ANOVA:
-
Analysis of variance
- BLASTn:
-
Basic local alignment search tool
- DMSO:
-
Dimethyl sulfoxide
- EI:
-
Electron impact ionization
- FID:
-
Flame ionization detector
- GC/MS:
-
Gas chromatography/mass spectrometry
- ITS:
-
Internal transcribed spacer region
- LSD:
-
Fisher's least significant difference
- NCBI:
-
National Centre for Biotechnology and Information
- PDA:
-
Potato dextrose agar
- PDB:
-
Potato dextrose broth
References
Adams RP (1995) 10. Identification of essential oil components by gas chromatography. Mass Spectrom:237–243
Akbar M, Javaid A (2015) Management of Rumex dentatus (toothed dock) by fungal metabolites under field conditions. Int J Agric Biol 17:187–192
Aybeke M (2017) Fusarium infection causes genotoxic disorders and antioxidant-based damages in Orobanche spp. Microbiol Res 201:46–51
Babu RM, Sajeena A, Seetharaman K (2003) Bioassay of the potentiality of Alternaria alternata (Fr.) Keissler as a bioherbicide to control waterhyacinth and other aquatic weeds. Crop Prot 22:1005–1013
Bajwa AA, Shabbir A, Adkins SW (2018) Interference and impact of parthenium weed on agriculture. Parthenium weed: Biol Ecol Manage 7:57
Bonito GM, Gryganskyi AP, Trappe JM, Vilgalys R (2010) A global meta-analysis of Tuber ITS rDNA sequences: species diversity, host associations and long-distance dispersal. Mol Ecol 19:4994–5008
Boulos L (1999) Flora of Egypt, vol 1. Al-Hadara Publishing, Cairo, Egypt, p 419
Boulos L (2002) Flora of Egypt, vol 3. Al-Hadara Publishing, Cairo, Egypt, p 373
Boulos L (2005) Flora of Egypt, vol 4. Al-Hadara Publishing, Cairo, Egypt, p 617
Boulos L (2009) Flora of Egypt Checklist. Revised Annotated Edition. Al-Hadara Publishing, Cairo, p 410
Cordeau S, Triolet M, Wayman S, Steinberg C, Guillemin JP (2016) Bioherbicides: dead in the water? A review of the existing products for integrated weed management. Crop Prot 87:44–49
de Souza ARC, Baldoni DB, Lima J, Porto V, Marcuz C, Ferraz RC, Kuhn RC, Jacques RJS, Guedes JVC, Mazutti MA (2015) Bioherbicide production by Diaporthe sp. isolated from the Brazilian Pampa biome. Biocatal Agric Biotechnol 4:575–578
Ekman R, Holmbom B (1989) Analysis by gas chromatography of the wood extractives in pulp and water samples from mechanical pulping of spruce. Nordic Pulp and Paper Res J (Sweden).
Guo QY, Cheng L, Zhu HX, Li W, Wei YH, Chen HY, Guo LZ, Weng H, Wang J (2020) Herbicidal activity of Aureobasidium pullulans PA-2 on weeds and optimization of its solid-state fermentation conditions. J Integr Agric 19:173–182
Javed S, Yousaf Z, Rashid M, Saleh N, Zahoor M, Ramzan H, Yasin H, Qamar NR, Aftab A (2016) Pericarp of Trapa natans var. bispinosa (Roxb.) Makino as an organic herbicide.
Júnior FWR, Scariot MA, Forte CT, Pandolfi L, Dil JM, Weirich S, Carezia C, Mulinari J, Mazutti MA, Fongaro G (2019) New perspectives for weeds control using autochthonous fungi with selective bioherbicide potential. Heliyon 5:e01676
Lee HB, Kim JC, Hong KS, Kim CJ (2008) Evaluation of a fungal strain, Myrothecium roridum F0252, as a bioherbicide agent. Plant Pathol J 24:453–460
Marley P, Shebayan J (2005) Field assessment of Fusarium oxysporum based mycoherbicide for control of Striga hermonthica in Nigeria. BioControl 50:389–399
Matthews J (2018) Management of herbicide resistant weed populations. Herbicide resistance in plants. CRC Press, In, pp 317–336
Mitchell JK (2003) Development of a submerged-liquid sporulation medium for the potential smartweed bioherbicide Septoria polygonorum. Biol Control 27:293–299
Mitchell JK, Yerkes CN, Racine SR, Lewis EH (2008) The interaction of two potential fungal bioherbicides and a sub-lethal rate of glyphosate for the control of shattercane. Biol Control 46:391–399
Mohankumar M, Vijayasamundeeswari A, Karthikeyan M, Mathiyazhagan S, Paranidharan V, Velazhahan R (2010) Analysis of molecular variability among isolates of Aspergillus flavus by PCR-RFLP of the ITS regions of rDNA. J Plant Prot Res 50:446–451
Moubasher AH (1993) Soil fungi in Qatar and other Arab countries. University of Qatar, The centre for scientific and applied research
Pes MP, Mazutti MA, Almeida TC, Curioletti LE, Melo AA, Guedes JV, Kuhn RC (2016) Bioherbicide based on Diaporthe sp. secondary metabolites in the control of three tough weeds. Afr J Agric Res 11:4242–4249
Sahin H (2019) A Review on parameters affecting the choice of alternative (non-chemical) weed control methods. Eur J Eng Res Sci 4:16–19
Saxena S, Pandey AK (2002) Evaluation of an indigenous isolate of Alternaria alternata (LC# 508) for use as a mycoherbicide for Lantana camara L. Crop Prot 21:71–73
Schütte G, Eckerstorfer M, Rastelli V, Reichenbecher W, Restrepo-Vassalli S, Ruohonen-Lehto M, Saucy AGW, Mertens M (2017) Herbicide resistance and biodiversity: agronomic and environmental aspects of genetically modified herbicide-resistant plants. Environ Sci Eur 29:5
Shabana YM (2005) The use of oil emulsions for improving the efficacy of Alternaria eichhorniae as a mycoherbicide for waterhyacinth (Eichhornia crassipes). Biol Control 32:78–89
Sica VP, Figueroa M, Raja HA, El-Elimat T, Darveaux BA, Pearce CJ, Oberlies NH (2016) Optimizing production and evaluating biosynthesis in situ of a herbicidal compound, mevalocidin, from Coniolariella sp. J Ind Microbiol Biotechnol 43:1149–1157
Täckholm V (1974) Students’ Flora of Egypt, 2nd edn. Cairo University, Giza, Egypt, p 888
Tuzi A, Andolfi A, Cimmino A, Evidente A (2010) X-ray crystal structure of phyllostin, a metabolite produced by Phyllosticta cirsii, a potential mycoherbicide of Cirsium arvense. J Chem Crystallogr 40:15
Varejão EVV, Demuner AJ, Barbosa LCDA, Barreto RW (2013) Phytotoxic effects of metabolites from Alternaria euphorbiicola against its host plant Euphorbia heterophylla. Quimica Nova 36:1004–1007
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18:315–322
Zhang W, Wolf TM, Bailey KL, Mortensen K, Boyetchko SM (2003) Screening of adjuvants for bioherbicide formulations with Colletotrichum spp. and Phoma spp. Biol Control 26:95–108
Zhang LH, Zhang JL, Liu YC, Cao ZY, Han JM, Juan Y, Dong JG (2013) Isolation and structural speculation of herbicide-active compounds from the metabolites of Pythium aphanidermatum. J Integr Agric 12: 1026-1032
Acknowledgements
The authors would like to extend their sincere appreciation to the Misr El Kheir Foundation: Science, Technology and Innovation (Cordeau et al.) Program to support this work by Lyophilizer instrument under the project code LGA05130114.
Funding
Not applicable
Author information
Authors and Affiliations
Contributions
All authors contributed equally to the work presented in this paper. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mohammed, Y.M.M., Badawy, M.E.I. Potential of phytopathogenic fungal isolates as a biocontrol agent against some weeds. Egypt J Biol Pest Control 30, 92 (2020). https://doi.org/10.1186/s41938-020-00295-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-020-00295-0