Abstract
Background
This study aimed to develop and assess a practical prognostic lncRNA signature for squamous cell carcinoma of the lung (LUSC).
Methods
RNA expression profile and clinical data from 388 LUSC patients were accessed and download from the Cancer Genome Atlas (TCGA) database. Differential lncRNA expression was compared and analyzed between normal tissue and tumor samples. By univariate and multivariate Cox regression analyses, a seven-lncRNA signature was developed and used for the purpose of survival prediction in LUSC patients. We applied receiver operating characteristic analysis to assess the performance of our model. The gene ontology enrichment analysis of seven lncRNA-related protein-coding genes was used to predict the potential biological functions of these lncRNAs.
Results
Sixteen out of 1414 differentially expressed lncRNAs in the TCGA dataset were associated with the overall survival of LUSC patients. Risk score analysis was used to select seven lncRNAs to be included in our model development and validation. The ROC analysis indicated that the specificity and sensitivity of this profile are high. Further functional enrichment analyses suggest that these lncRNAs may regulate genes that affect the function of the major histocompatibility complex and the cell membrane.
Conclusions
The current study identified a seven-lncRNA signature that predicts the outcome of LUSC, offering potentially novel therapeutic targets for the treatment of squamous cell carcinoma of the lung.
Similar content being viewed by others
Background
Lung cancer remains the leading cause of cancer death in the world [1], in which non-small cell lung cancer (NSCLC) is the most frequent type of lung cancer, including adenocarcinomas (LAD), squamous cell carcinomas (LUSC) and large cell carcinoma (LCC) [2]. LUSC represents a major public health issue, accounting for 27% of all lung cancers. LUSC exhibits distinct epidemiological, clinicopathological and molecular characteristics [3]. However, effective biomarkers for early detection, prediction of high recurrence populations and risk of death and the identification of target therapies are still lacking. Thus, identification of effective biomarkers for the prognosis of LUSC is critical for the diagnosis and treatment of LUSC patients.
Long non-coding RNAs (lncRNA) regulate gene transcription and are implicated in diverse biological processes. With the importance of lncRNAs being investigated in cancer research, the alterations of the lncRNA landscape [4] and roles of lncRNAs as drivers of tumor suppression and oncogenesis have been identified [5]. Moreover, long non-coding RNAs in circulation have been found in patient blood samples and act as a novel biomarker in plasma for predicting NSCLC [6, 7]. This suggests that lncRNAs may be non-invasive biomarkers for lung cancer.
Although a number of lncRNAs have been identified for predicting the outcomes in NSCLC [5], the prognostic value of a single candidate lncRNA biomarker is limited. This may be due to the small sample sizes as well as inconsistent sample collection and detection methods in previous studies. Identifying lncRNA expression signatures that are associated with patient survival in standard clinical samples may lead to the discovery of molecular drug subclasses and potential drug targets. Several prognostic gene expression signatures have been published for NSCLC [8,9,10], but none of these studies includes lncRNAs in a large cohort to identify and assess the prognostic value of lncRNA biomarkers for LUSC patients. Moreover, the molecular characteristics [11] and prognosis pattern differ between LUSC and LUAD, and we focused on the lncRNA survival signature of LUSC not previously reported.
We applied a survival associated risk-score formula to identify a novel 7-lncRNA prognostic signature from the TCGA dataset of 388 LUSC patient samples. To show the robustness of this signature, the specificity and sensitivity of our model was tested by the area under ROC curve (AUROC) analysis.
Methods
Datasets
LncRNA RNA-seq data (HTSeq-FPKM-UQ) comprised of 504 LUSC patients was obtained from the publicly available Genomic Data Commons (GDC) data portal (https://portal.gdc.cancer.gov/). Corresponding clinical data, including age, gender, smoking history and TNM stage were also obtained and assessed. We excluded incomplete clinical data or overall survival (OS) of less than 1 month in this analysis. After exclusions, a total of 388 LUSC patients were enrolled in the development of our model. The 388 LUSC patients were randomly divided into a training set (n = 194) and a testing set (n = 194). Patient IDs in both training and testing sets are shown in Additional file 1: Table S1. The training set was used to identify the lncRNA expression signature, and the testing set was used for further validation.
Identification of differentially expressed lncRNAs in LUSC
All analyses were performed using R version 3.3.0. To identify lncRNAs suitable for subsequent survival analyses, we utilized the trimmed mean of M values method for normalization and differential expression analysis using the edgeR package from Bioconductor [12, 13]. The parameters for screening the expression difference of lncRNAs were padj <0.01 and |log2FoldChange| > 2.
Cox regression analysis
First, the RNA-seq expression values were transformed (log2) to normalize the data. The association between lncRNA expression and patient survival was determined by univariate Cox regression analysis using the Survival R package from CRAN [14]. The lncRNAs (p-value < 0.01) from the univariate analysis were used to mine potential candidate lncRNAs associated with OS. The Cox proportional hazard model was applied for multivariate analysis to identify covariates with independent prognostic value. The best mathematical model was built based on the Akaike Information Criterion (AIC) [15], which allowed for the determination of the best trade-off between the complexity of a model and its goodness of fit.
Risk score and survival curve
A mathematical formula (Risk score = 0.052*LINC01412 - 0.047*RP11-277P12.9 - 0.051*RP11-60H5.1 + 0.066*RP11-697M17.2 + 0.034*RP11-897M7.1 + 0.050*CTB-43E15.2 + 0.036*RP11-0.036*H4.1) was developed to predict the risk score for each patient based on the multivariate Cox regression analysis. According to our risk scoring system, patients were divided into low-risk and high-risk groups according to the median risk score. Subsequently, the log-rank test was used to determine the differences in survival. A Kaplan-Meier overall survival curve of the two groups was plotted and the hazard ratio was calculated. Cox multivariate analysis was also used to test whether or not the risk score was independent of the clinical parameters, such as age, gender, smoking history and tumor stage. The prognostic performance was measured using the Survival ROC R package from CRAN [16].
In silico functional pathways analysis
We examined the correlation between the expression level of the seven lncRNAs and each protein coding gene (PCGs) using two-sided Pearson correlation coefficients and the z-test [17]. The PCGs positively or negatively correlated with the seven lncRNAs were considered as lncRNA-related PCGs (|Pearson correlation coefficient| > 0.4 and P-value < 0.01). Gene ontology (GO) enrichment analysis of lncRNA-related PCGs was analyzed by using the DAVID online tool (https://david.ncifcrf.gov/) [18]. The GO terms with P-values of <0.05 were considered as significantly enriched functions of prognostic lncRNAs. Significant GO terms with similar functions were organized into an interaction network and visualized using the Enrichment Map plugin for Cytoscape 3.2.1 (http:// baderlab.org/Software/EnrichmentMap/) [19].
Results
Patient characteristics
According to the defined criteria, a total of 388 LUSC patients with both RNA-seq expression profiles and clinical data [20] were downloaded from the GDC data portal. The Clinical covariates of the patients and tumors in both training and test sets are showed in Table 1. Of the 388 patients, 183 had Stage I disease, 130 had Stage II, 69 were labeled with Stage III and 6 with Stage IV disease. For subsequent model development, we randomly divided all the patients into the training set (n = 194) and testing set (n = 194) as previously reported [21, 22]. There was no significant difference in the clinical covariates between the two sets (P > 0.05) (Table 1).
Differentially expressed lncRNAs in LUSC patients
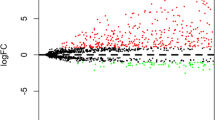
A total of 1414 lncRNAs were found to be differentially expressed between LUSC and normal lung tissues, and were used for survival analyses (Additional file 2: Table S2). To identify the lncRNAs which are associated with patient survival in LUSC, univariate Cox regression analysis for all lncRNA expression data was assessed [23]. With the significance level cutoff threshold of 0.01, a set of 16 lncRNAs were selected (Additional file 3: Table S3). These lncRNAs were used in stepwise multivariate Cox regression analysis and, finally, seven lncRNAs (LINC01412, RP11-277P12.9, RP11-60H5.1, RP11-697 M17.2, RP11-897M7.1, CTB-43E15.2 and RP11-366H4.1) were identified (Fig. 1). We conducted a risk score analysis of the seven lncRNAs to calculate the risk score for each patient [24]. The risk score formula for our model is listed in Table 2 (Risk score = 0.052*LINC01412 - 0.047*RP11-277P12.9 - 0.051*RP11-60H5.1 + 0.066*RP11-697M17.2 + 0.034*RP11-897M7.1 + 0.050*CTB-43E15.2 + 0.036*RP11-366H4.1). Of these seven lncRNAs, five were associated with high risk (LINC01412, RP11-697M17.2, RP11-897M7.1, CTB-43E15.2, RP11-366H4.1, Coef > 0) and two were shown to be protective (RP11-277P12.9, RP11-60H5.1, Coef < 0) (Fig. 1).
The development of the 7-lncRNA prognostic model
We divided the patients into high-risk and low-risk groups according to the median risk score (value = 0.909) calculated from the expression levels of the seven lncRNAs. The log-rank test was used to determine the survival differences. As depicted in Fig. 2a, Kaplan-Meier curves indicated that the high-risk group was correlated with poor prognosis in the training set (p < 0.0001). ROC curves indicated that the AUC of the 7-lncRNA signature was 0.694 in the training set (Fig. 2b), which showed that the 7-lncRNA signature had a high specificity and sensitivity in predicting the overall survival time of LUSC patients.
Kaplan-Meier and ROC curves for the 7-lncRNA signature in the training set. a The differences between the high-risk (n = 97) and low-risk (n = 97) groups were determined by the log-rank test (p < 0.0001). Five year overall survival was 36.4% (95% CI: 25.5%-52.1%) and 65.3% (95% CI: 53.7%-79.4%) for the high-risk and low-risk groups, respectively. b ROC curves indicated that the area under receiver operating characteristic of 7-lncRNA model was 0.694
In order to validate the prognostic power of the 7-lncRNA model, the log-rank statistical test was performed in the testing set. Patients in the validation set were divided into low-risk and high-risk groups according to the previous median risk score of the training set (value = 0.909). As in the training set, statistically significant differences (P<0.05) between the low-risk group and the high-risk group were observed (Fig. 3a),indicating that our 7-lncRNA signature is suitable for the prediction of LUSC patient survival.
Kaplan-Meier and ROC curves for the 7-lncRNA signature in the validation set. a The differences between the high-risk (n = 103) and low-risk (n = 91) groups were determined by the log-rank test (p < 0.0001). Five year overall survival was 36.8% (95% CI: 26.1%-51.8%) and 61.9% (95% CI: 51.4%-74.6%) for the high-risk and low-risk groups, respectively. b ROC curves indicated that the area under receiver operating characteristic of 7-lncRNA model was 0.685
To verify whether or not the 7-lncRNA model could distinguish the risk from the pool of all LUSC patients when potential prognostic factors were taken into account, a multivariate analysis was performed to evaluate the independent prognostic value of the model. Among the demographic data associated with the prognosis of cancer, the results indicated that the 7-lncRNA signature served as a strong independent predictor of LUSC overall survival (high-risk, HR: 2.822, 95% CI 2.026–3.929, p < 0.0001, Table 3), compared with clinical data such as age, gender and TNM stage.
Functional enrichment analysis of pathways correlated with the prognostic lncRNAs in LUSC
After the measurement of the correlation of the lncRNAs in our model and those of the PCGs, co-expression between 444 genes and at least one of the seven lncRNAs (|Pearson correlation coefficient| > 0.4 and P-value < 0.01) was found. The 444 PCGs clustered most significantly in the GO enrichment (Additional file 4: Table S4) categories major histocompatibility complex (GO:0042613, GO:0032395, GO:0023026) and membrane (GO:0005886, GO:0016021, GO:0009897, GO:0030666) in our analysis (Fig. 4). These results suggest that the lncRNAs of the signature may regulate genes that affect the adaptive immune system and the function of the cell membrane.
Discussion
Increasing evidence reveals that lncRNAs play crucial roles in the tumorigenesis and progression of lung cancer. Although several studies have identified a number of lncRNAs with prognostic value in NSCLC, no studies have focused on and analyzed the expression of lncRNAs in LUSC. Moreover, because LUSC has distinct molecular characteristics [11], single lncRNA expression patterns are not sufficient for accurate prediction of LUSC outcomes. Therefore, we focused on the prognostic lncRNA expression patterns in lung squamous cell carcinoma.
In the current study, 7 of the 1414 differentially expressed lncRNAs associated with overall survival of LUSC patients were identified. Using univariate Cox regression analysis and stepwise multivariate Cox regression analysis, a novel seven-lncRNA (LINC01412, RP11-277P12.9, RP11-60H5.1, RP11-697M17.2, RP11-897M7.1, CTB-43E15.2, RP11-366H4.1) signature was established and validated to demonstrate high specificity and sensitivity in predicting the overall survival time of LUSC patients.
In order to gain a further insight into the functional roles of the seven lncRNAs, the correlation between their expression levels and the co-expressed protein coding genes was analyzed. Bioinformatic analysis revealed that 444 co-expressed protein coding genes clustered most significantly in the major histocompatibility complex (MHC) and membrane proteins in GO enrichment categories (Additional file 4: Table S4). MHC molecules exert their role in the immunological recognition and participate in destruction of tumor cells. Végh et al. reported that the loss frequency of MHC class I molecules was 36% (5 of 14 cases) in primary lung carcinomas [25]. In addition, the loss of MHC class I and MHC-encoded transporter TAP-1, which is necessary in antigenic peptide transportation, has been observed frequently in lung cancer, although no relationship between the loss of these molecules and patient survival was determined [26]. It is possible this relationship was not found due to the small sample size employed in the study. Recently, MHC II NSCLC vaccines have been reported as potential immunotherapies for a range of NSCLC patients, including LUSC [27]. Passlick et al. found that immunologically relevant cell surface molecules are frequently expressed in primary NSCLC, which is consistent with our results. However, no evidence showed how MHC molecules impacted the course of cancer [28]. Since the MHC and membrane proteins play an important role in vaccine and immune therapy target design, understanding how lncRNAs epigenetically regulate adaptive immune function through MHC and membrane proteins, subsequently affecting LUSC survival, is crucial.
Conclusion
In summary, our study identified a novel seven-lncRNA prognostic signature as a specific predictor for LUSC patients. In addition to TNM staging and qualified sampling methods to avoid bias and intratumor heterogeneity, further molecular investigations, such as exploring the underlying mechanisms of these lncRNAs in LUSC development and using independent cohorts of large sample sizes from multiple institutions, are necessary in order to confirm these predictions.
Abbreviations
- AIC:
-
The Akaike Information Criterion
- AUROC:
-
Area under the receiver operating characteristic curve
- GO:
-
Gene Ontology
- lncRNA:
-
Long non-coding RNA
- LUSC:
-
Lung Squamous Cell Carcinoma
- MHC:
-
Major histocompatibility complex
- NSCLC:
-
Non-small cell lung cancer
- PCGs:
-
Protein-coding genes
- RNA-seq:
-
RNA Sequencing
- TCGA:
-
The Cancer Genome Atlas
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30.
Feinstein MB, Bach PB. Epidemiology of lung cancer. Chest Surg Clin N Am. 2000;10(4):653–61.
Drilon A, Rekhtman N, Ladanyi M, Paik P. Squamous-cell carcinomas of the lung: emerging biology, controversies, and the promise of targeted therapy. Lancet Oncol. 2012;13(10):e418–26.
White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 2014;15(8):429.
Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407.
Tang Q, Ni Z, Cheng Z, Xu J, Yu H, Yin P. Three circulating long non-coding RNAs act as biomarkers for predicting NSCLC. Cell Physiol Biochem. 2015;37(3):1002–9.
Peng H, Wang J, Li J, Zhao M, Huang SK, Gu YY, Li Y, Sun XJ, Yang L, Luo Q, et al. A circulating non-coding RNA panel as an early detection predictor of non-small cell lung cancer. Life Sci. 2016;151:235–42.
Zhu CQ, Strumpf D, Li CY, Li Q, Liu N, Der S, Shepherd FA, Tsao MS, Jurisica I. Prognostic gene expression signature for squamous cell carcinoma of lung. Clin Cancer Res. 2010;16(20):5038–47.
Larsen JE, Pavey SJ, Passmore LH, Bowman R, Clarke BE, Hayward NK, Fong KM. Expression profiling defines a recurrence signature in lung squamous cell carcinoma. Carcinogenesis. 2007;28(3):760–6.
Raponi M, Zhang Y, Yu J, Chen G, Lee G, Taylor JM, Macdonald J, Thomas D, Moskaluk C, Wang Y, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66(15):7466–72.
Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–25.
Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25.
Robinson MD, McCarthy DJ. Smyth GK: edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40.
Therneau T, Lumley T. Survival: Survival analysis, including penalised likelihood. R package version 2.36-5. Survival: Survival analysis, including penalised likelihood R package version 2011:2.36-32.2010. https://rweb.stat.umn.edu/R/site-library/survival/html/00Index.html.
Bozdogan H. Model selection and Akaike's information criterion (AIC): the general theory and its analytical extensions. Psychometrika. 1987;52(3):345–70.
Heagerty PJ: survivalROC: Time-dependent ROC curve estimation from censored survival data. 1923.
Bhardwaj N, Lu H. Correlation between gene expression profiles and protein--protein interactions within and across genomes. Bioinformatics. 2005;21(11):2730–8.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocol. 2009;4(1):44–57.
Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5(11):e13984.
Lee H, Palm J, Grimes SM, Ji HP. The cancer genome atlas clinical explorer: a web and mobile interface for identifying clinical-genomic driver associations. Genome Med. 2015;7:112.
Shi H, Chen J, Li Y, Li G, Zhong R, Meng R, Kong W, Lu M. Identification of a six microRNA signature as a novel potential prognostic biomarker in patients with head and neck squamous cell carcinoma. Oncotarget 2016;7(16):21579–90.
Li X, Shi Y, Yin Z, Xue X, Zhou B. An eight-miRNA signature as a potential biomarker for predicting survival in lung adenocarcinoma. J Transl Med. 2014;12(1):1.
Zhou M, Zhao H, Wang Z, Cheng L, Yang L, Shi H, Yang H, Sun J. Identification and validation of potential prognostic lncRNA biomarkers for predicting survival in patients with multiple myeloma. J Exp Clin Cancer Res. 2015;34(1):1.
Li J, Chen Z, Tian L, Zhou C, He MY, Gao Y, Wang S, Zhou F, Shi S, Feng X. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut 2014;63(11):1700-10. doi:10.1136/gutjnl-2013-305806. Epub 2014 Feb 12.
Vegh Z, Wang P, Vanky F, Klein E. Selectively down-regulated expression of major histocompatibility complex class I alleles in human solid tumors. Cancer Res. 1993;53(10 Suppl):2416–20.
Korkolopoulou P, Kaklamanis L, Pezzella F, Harris AL, Gatter KC. Loss of antigen-presenting molecules (MHC class I and TAP-1) in lung cancer. Br J Cancer. 1996;73(2):148–53.
Srivastava MK, Bosch JJ, Wilson AL, Edelman MJ, Ostrand-Rosenberg S. MHC II lung cancer vaccines prime and boost tumor-specific CD4+ T cells that cross-react with multiple histologic subtypes of nonsmall cell lung cancer cells. Int J Cancer. 2010;127(11):2612–21.
Passlick B, Izbicki JR, Simmel S, Kubuschok B, Karg O, Habekost M, Thetter O, Schweiberer L, Pantel K. Expression of major histocompatibility class I and class II antigens and intercellular adhesion molecule-1 on operable non-small cell lung carcinomas: frequency and prognostic significance. Eur J Cancer. 1994;30A(3):376–81.
Acknowledgments
We would like to extend our sincere gratitude to Mark Deiparine for manuscript editing services.
Availability of data and materials
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Funding
This work was supported by the Science and Technology Planning Project of Guangdong Province, China (411234349027), Clinical Application and Translational Medicine Research Project of the First Affiliated Hospital of Guangzhou University (201515-gyfyy) and the Open Project Program of the State Key Laboratory of Respiratory Disease (2014SKLRD-O09).
Author information
Authors and Affiliations
Contributions
Xiaobing Le, Kailing Huang, Viola Yingjun Luo, Yanhui Liu and Allen M. Chen downloaded and analyzed the TCGA data and performed the bioinformatic analysis. Xiaoshun Shi, Fuxi Huang, Xiaoxiang Li, Ying Liang and Jiexia Zhang analyzed and interpreted the patient data reported in the TCGA data and evaluated their clinical significance. Jiexia Zhang,Ying Liang, Allen M. Chen and Xiaoshun Shi conceived and designed the study. Xiaoshun Shi and Fuxi Huang were major contributors in writing and revising the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
None required, as all data were downloaded from internet databases that have ethics approvals included with previously published materials.
Consent for publication
None.
Competing interests
The authors have declared that no conflicts of interest exist. There are no potential conflicts with Kailing Huang, Viola Yingjun Luo,Yanhui Liu, Allen M. Chen or Mendel Genes, Inc.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1: Table S1.
Patient IDs in the training and testing sets. (XLSX 13 kb)
Additional file 2: Table S2.
Differentially expressed lncRNAs. (XLSX 92 kb)
Additional file 3: Table S3.
Univariate Cox regression result. (XLSX 10 kb)
Additional file 4: Table S4.
Gene Ontology (GO) Enrichment Analysis. (XLSX 39 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Shi, X., Huang, F., Le, X. et al. A practical prognostic lncRNA signature for lung squamous cell carcinoma. transl med commun 2, 7 (2017). https://doi.org/10.1186/s41231-017-0016-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41231-017-0016-6