Abstract
Background
Lung squamous cell carcinoma (LSCC) is a form of cancer that is associated with high rates of relapse, poor responsiveness to therapy, and a relatively poor prognosis. The relationship between long non-coding RNA (lncRNA) expression and LSCC patient prognosis remains to be established.
Methods
In the present study, we discovered that lncRNAs were differentially expressed in LSCC tumor tissues relative to normal control tissues, and we explored the prognostic relevance of these lncRNA expression patterns using data from the Cancer Genome Atlas (TCGA).
Results
These multidimensional data were analyzed in order to identify lncRNA signatures that were associated with LSCC patient survival outcomes. Kaplan-Meier survival curves revealed prognostic capabilities for three of these lncRNAs (LINC02555, APCDD1L-DT and OTX2-AS1). A Cox regression analysis revealed this three-lncRNA signature to be significantly associated with patient survival. Further GO and KEGG analyses revealed that the predicted target genes of these three lncRNAs were also potentially involved in cancer-associated pathways.
Conclusions
Together these results thus indicate that this novel three-lncRNA signature can be used to predict LSCC patient prognosis.
Similar content being viewed by others
Background
Lung cancer is a highly heterogeneous disease, with genetic, epigenetic, and environmental factors all acting to shape its development and progression. Lung cancer mortality rates are the highest of all forms of cancer, accounting for 25 and 30% of all cancer-associated deaths in the USA and China, respectively [1, 2]. In 2015 alone, 733,000 new cases of lung cancer were diagnosed in China (69% in males and 31% in females), while 218,527 new cases were diagnosed in the USA during this same period (52% in males and 48% in females). SEER data indicate that lung cancer patients exhibit a 5-year survival rate of just 18.1% [3]. Lung squamous cell carcinoma (LSCC) cases account for a significant fraction of overall lung cancer cases [4]. LSCC more often occurs in men, is related to the smoking of tobacco, and is often associated with high rates of relapse, poor responsiveness to therapeutic intervention, and a generally poor patient prognosis [5, 6]. While there have been many advances in the field of clinical oncology as a whole in recent years, rates of 5-year overall survival (OS) for LSCC patients still remain low. As such, it is vital that novel approaches be identified that can be used to predict the prognosis of LSCC patients so as to guide clinical decision making and treatment efforts in these individuals.
Long non-coding RNAs (lncRNAs) are RNA molecules > 200 nucleotides in length that lack coding potential [7]. Emerging evidence indicates that some lncRNAs do encode proteins and play roles in transcriptional, and epigenetic gene regulation, and cancer [8, 9]. These lncRNAs have been shown to frequently be dysregulated in cancer, with their altered expression patterns having a direct impact on tumor cell gene expression at the post-transcriptional and epigenetic levels, as well as on the proliferation, survival, invasion, and metastasis of these cells [10,11,12]. However, relatively few studies to date have specifically examined the relationship between lncRNA expression and LSCC patient prognosis. In the present study, we therefore explored patterns of differential lncRNA expression in LSCC tumor tissues and normal control tissue samples in an effort to assess the prognostic relevance of such lncRNA expression patterns. Through this approach we were able to develop a three lncRNA signature which was found to be significantly associated with the survival of LSCC patients.
Materials and methods
LSCC patient datasets
Level 3 expression and clinical data pertaining to 409 LSCC and 49 control samples were downloaded from The Cancer Genome Atlas (TCGA, https://tvga-data.nci.nih.gov/tcga/). Datasets and patient records were used in order to assess both patterns of lncRNA expression as well as clinocpathological and demographic variables including gender, age at time of diagnosis, and TNM staging (Table 1). The mean follow-up time was 3.5 years. The Ethics Committee of the Institutional Review Board of Ningbo Yinzhou Second Hospital and Cixi People’s Hospital in Zhejiang Province approved this study. Samples were included in the present analysis if they were from patients with an OS > 1 month for whom lncRNA differential expression data and information pertaining to clinical details and prognosis were available. The language package in R was used in order to interpret the lncRNA sequencing data, while the limma package was used when assessing differential lncRNA expression between LSCC and control samples, with differential expression being expressed based upon fold change (FC) values. Those lncRNAs with a log2|FC| > 1.0 and p < 0.05 were considered to be significantly differentially expressed.
Statistical analysis
The prognostic relevance of differentially expressed lncRNAs in LSCC was assessed using Kaplan-Meier curves and log-rank tests. We ultimately constructed a signature using a linear combination of the expression levels of these three lncRNAs and the estimated regression coefficients in the multivariable Cox regression analysis. A mathematical formula (Risk score = 0.0768*LINC02555 + 0.0917* APCDD1L-DT – 0.1176*OTX2-AS1) was developed to predict the risk score for each patient based on the multivariable Cox regression analysis. This three lncRNA signature-derived risk score was then used to stratify patients into high- and low-risk groups, using the median risk score in this cohort as a cutoff point for stratification purposes. Kaplan-Meier curves and log-rank tests were then used to compare survival outcomes between these high- and low-risk patients. In addition, receiver operating characteristic (ROC) analyses were used in order to compare the sensitivity and specificity of this three lncRNA risk score as a means of predicting patient survival outcomes. P < 0.05 was the significance threshold. R version 3.5.1 [13] was used for all statistical testing.
Functional analysis
Correlating genes to the differentially expressed lncRNAs were obtained using the co-expression method. Pearson correlation coefficients between the expression profiles of the three prognostic lncRNAs and their protein-coding genes (PCGs) were calculated to determine their relationships. Those PCGs with a Pearson’s R > 0.40 and p < 0.05 were considered to be lncRNA-related. These putative lncRNA targets were then subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analyses. Furthermore, these target genes were incorporated into a protein-protein interaction (PPI) network using the STRING database [14], with Cytoscape being used for network visualization [15]. Select protein pairs from this network with > 10 nodes were the outputs of this analysis.
Results
Patient characteristics
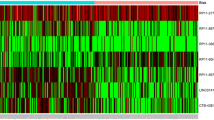
The study investigated 458 patient samples. The mean follow-up time was 3.5 years and 200 patient samples were dead in this study. Table 1 lists detailed clinical characteristics, including gender, race, age at diagnosis, and disease stage. Of the enrolled patients, 26.6% were female, and 78.6% were older than 60 years. The most common tumor grades were I (48.9%) and II (32.8%). A total of 936 differentially expressed lncRNAs, including 687 upregulated and 249 downregulated lncRNAs, were identified between LSCC and normal tissues in Fig. 1.
The relationship between lncRNA expression and LSCC patient OS
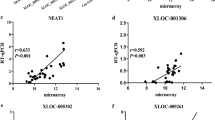
We began by using univariable and multivariable Cox proportional hazard regression models in order to identify those lncRNAs which were associated with LSCC patient prognosis. In total, we identified three candidate lncRNAs in these LSCC patients in univariable Cox model (p < 0.01; Fig. 2a). A multivariable model confirmed that the expression of the lncRNAs LINC02555 (HR = 1.136, p < 0.001), APCDD1L-DT (HR = 1.136, p < 0.001), and OTX2-AS1 (HR = 0.859, p < 0.001) were all independently associated with LSCC patient OS (Fig. 2b). Kaplan-Meier survival curves and log-rank tests were further used to examine the relationship between these lncRNAs and patient survival. We found that two of the tested lncRNAs (LINC02555 and APCDD1L-DT) were negatively associated with LSCC patient OS, whereas the lncRNA OTX2-AS1 was positively correlated with OS (Fig. 3).
Kaplan-Meier method and log-rank test revealed that three lncRNAs were associated with OS in patients with LSCC. The patients were divided into low and high expression levels group according to the median value. a APCDD1L-DT b LINC02555 and (C) OTX2-AS1: a population-based study with a mean follow-up of 3.5 years
The prognostic utility of the three lncRNA signature
Using this three lncRNA signature, we were able to assign risk scores to patient samples, after which these samples were separated into high- and low-risk groups based upon the median risk score value. We then found that patients in the high-risk group had a significantly shorter OS than did patients in the low-risk group (p < 0.001) (Fig. 4a). We then used an ROC analysis in order to assess the prognostic utility of this three lncRNA signature. The AUC values for these curves as predictors of LSCC patient 3- and 5-year survival were 0.675 and 0.613, respectively, corresponding to an effective survival prediction (Fig. 4b). Patients in the high-risk group expressed higher levels of the lncRNAs LINC02555 and APCDD1L-DT on average relative to low risk patients, whereas low-risk patients expressed higher levels of OTX2-AS1 lncRNA.
Functional enrichment analysis and PPI networks
In order to identify potential targets for these three lncRNAs which were associated with LSCC patient prognosis, we conducted a co-expression analysis as detailed in the Materials and Methods section. We then performed GO and KEGG pathway analyses on these co-expressed genes in order to unravel their potential physiological roles (Fig. 5). These co-expressed genes were primarily enriched in genes associated with biological processes such cell adhesion molecule binding, ubiquitin-like protein ligase binding, protein serine/threonine kinase activity, ubiquitin protein ligase binding, actin binding, ATPase activity, phospholipid binding, and phosphoric ester hydrolase activity. These genes were additionally significantly enriched in KEGG pathways including endocytosis, focal adhesion, MAPK signaling, lysosomes, neurotrophin signaling, ubiquitin mediated proteolysis, axon guidance, herpes simplex virus 1 infection, and the cell cycle. Furthermore, PPI networks were also obtained using the STRING tool. As mentioned in Fig. 5c, INS, GCG, GCGR, GLP1R, IAPP, P2RY1, CRH and FFAR1 had the most connections with other members of the module and were thus the most noteworthy nodes in this network.
Discussions
Lung cancer currently ranks as the deadliest form of cancer, and as such it is a primary focus for many cancer research efforts [16]. While LSCC patient prognosis has improved significantly in recent years owing to improvements in multidisciplinary treatment strategies, and chemotherapeutic/radiotherapeutic treatment regimens, LSCC recurrence rates remain high and as such this disease can impose a heavy burden upon patients, their families, and on medical institutions [17, 18]. Difficulties in accurately diagnosing LSCC and in predicting patient outcomes have led to low 5-year survival rates in affected patients [2]. As such, it is vital that novel biomarkers that can reliably predict LSCC patient outcomes be identified. It is similarly important that the molecular mechanisms governing the development and progression of LSCC be fully elucidated.
Many studies have clearly shown that the development of LSCC can be driven by interactions between genetic, transcriptomic, and proteomic factors [19, 20]. Changes in lncRNA expression patterns can also influence all stages of the oncogenic process, yet the prognostic relevance of these lncRNAs has not been sufficiently studied to date. As such, in the present study we examined lncRNA expression patterns in LSCC and were thus able to identify three lncRNAs that were significantly linked with LSCC patient OS. These three lncRNAs were then subjected to additional analyses aimed at identifying their putative target genes and potential biological roles through the use of pathway enrichment analyses. These results indicated that these three lncRNAs may play roles in regulating LSCC molecular pathogenesis, clinical progression, and patient prognosis, thus clearly demonstrating the prognostic relevance of lncRNA expression patterns in LSCC patients in a clinical setting.
Multiple studies [21, 22] have demonstrated that functional lncRNA expression can modulate oncognesis via altered regulation of gene expression and signaling within tumor cells. Indeed, certain lncRNAs are able to promote the development, progression, and metastasis of tumors through their ability to regulate the proliferation, differentiation, migration, and survival of these cancerous cells [22]. Huang et al. [23] found that increasing the expression of the downregulated lncRNA LINC00961 resulted in increased Bax expression and the corresponding apoptotic death of NSCLC cells. Xu et al. further provided evidence suggesting that the lncRNA HULC is able to promote LSCC cell proliferation owing to its ability to PTPRO-dependent phosphorylation and activation of NF-κB [24]. Similarly, Wang et al. found that increased expression of the lncRNA MIR31HG in NSCLC led to enhanced tumor cell gefitinib resistance owing to associated activation of the EGFR/PI3K/AKT signaling pathway [25]. Li J et al. reported lncRNA-ATB overexpression may promote the progression of LSCC by modulating the microRNA-590-5p/NF-90 axis [26]. To improve the prediction accuracy, we analyzed high-throughput data and were thereby able to identify two upregulated lncRNAs (LINC02555 and APCDD1L-DT) and one downregulated lncRNA (OTX2-AS1) in LSCC patients, all three of which were significantly associated with patient clinical outcomes.
Up to now, some researches have demonstrated that cigarette smoking or exposure may be associated with the expression of lncRNAs in lung cancer patients. Li J et al. [27] found that polymorphisms in lncRNA AC016683.6 significantly increased the risk of lung cancer in the smoking population. Lv X et al. also reported that there were significant interactions of lncRNA AC008392.1 polymorphisms with smoking exposure to lung cancer susceptibility [28]. Moreover, Chen Y et al. suggested that smoking-associated lncRNAs have a role in various processes and pathways, including cell proliferation and the cyclic guanosine monophosphate cGMP)/protein kinase cGMP-dependent 1 signaling pathway via bioinformatics analysis [29]. However, the association between cigarette smoking exposure and the identified lncRNAs (LINC02555, APCDD1L-DT and OTX2-AS1) in present study has not been investigated. Further studies are needed to confirm these predictions.
We further sought to gain insight into the functional importance of the three lncRNAs identified in this study via using a co-expression analysis-based approach to identify putative lncRNA target genes that were then subjected to GO and KEGG enrichment analyses. This approach revealed the lncRNA-associated target genes to be enriched for functionality in the context of endocytosis, focal adhesion, MAPK signaling, and lysosomal activity, all of which are closely linked with oncogenesis and tumor progression [30, 31]. To date no studies have specifically studied LINC02555, APCDD1L-DT, or OTX2-AS1 in the context of LSCC. As such, future in-depth molecular analyses will be needed to confirm the findings of our co-expression analysis.
There are multiple limitations to the present study. For one, these results are derived solely from bioinformatics analyses and as such necessitate additional functional validation. Furthermore, we did not explore the molecular mechanisms linking the expression of these three lncRNAs to LSCC patient prognosis, and as such future experimental studies will be required in order to elucidate these mechanisms. As such, large-scale multi-center trials will be essential in order to validate and expand upon our findings.
Conclusions
In summary, in the present article we were able to identify three different lncRNAs that could be used to predict survival outcomes in patients with LSCC. Further large-scale multi-center trials will be needed to confirm our findings, and to explore the molecular mechanisms linking these lncRNAs to clinical outcomes in LSCC patients. While much work is still required before this lncRNA signature can be implemented in a clinical setting, we nonetheless feel that our findings may have significant value as a future diagnostic or prognostic tool in the context of LSCC patient identification and care.
Abbreviations
- LSCC:
-
Lung squamous cell carcinoma
- LncRNAs:
-
Long non-coding RNAs
- TCGA:
-
The Cancer Genome Atlas
- PCGs:
-
Protein-coding genes
- ROC:
-
Receiver operating characteristic
- OS:
-
Overall survival
- FCs:
-
Fold changes
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- GO:
-
Gene Ontology
- BP:
-
Biological process
- NF-κB:
-
Nuclear factor-κB
References
American Thoracic Society/European Respiratory Society International Multidisciplinary Consensus Classification of the Idiopathic Interstitial Pneumonias. This joint statement of the American Thoracic Society (ATS), and the European Respiratory Society (ERS) was adopted by the ATS board of directors, June 2001 and by the ERS executive committee, June 2001. Am J Respir Crit Care Med. 2002;165(2):277–304.
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32. https://doi.org/10.3322/caac.21338.
Mayer JE, Swetter SM, Fu T, Geller AC. Screening, early detection, education, and trends for melanoma: current status (2007–2013) and future directions: Part II. Screening, education, and future directions. J Am Acad Dermatol. 2014;71(4):611.e1–611.e10 quiz 621-2.
Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6(2):244–85. https://doi.org/10.1097/JTO.0b013e318206a221.
Kenfield SA, Wei EK, Stampfer MJ, Rosner BA, Colditz GA. Comparison of aspects of smoking among the four histological types of lung cancer. Tob Control. 2008;17(3):198–204. https://doi.org/10.1136/tc.2007.022582.
Antal CE, Hudson AM, Kang E, Zanca C, Wirth C, Stephenson NL, et al. Cancer-associated protein kinase C mutations reveal kinase's role as tumor suppressor. Cell. 2015;160(3):489–502. https://doi.org/10.1016/j.cell.2015.01.001.
Carpenter S. Long noncoding RNA: novel links between gene expression and innate immunity. Virus Res. 2016;212:137–45. https://doi.org/10.1016/j.virusres.2015.08.019.
Li CH, Chen Y. Insight into the role of long noncoding RNA in cancer development and progression. Int Rev Cell Mol Biol. 2016;326:33–65. https://doi.org/10.1016/bs.ircmb.2016.04.001.
Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15(6):423–37. https://doi.org/10.1038/nrg3722.
Zhang B, Dong Y, Zhao Z. LncRNA MEG8 regulates vascular smooth muscle cell proliferation, migration and apoptosis by targeting PPARα. Biochem Biophys Res Commun. 2019;510(1):171–6. https://doi.org/10.1016/j.bbrc.2019.01.074.
Huang Y, Ni R, Wang J, Liu Y. Knockdown of lncRNA DLX6-AS1 inhibits cell proliferation, migration and invasion while promotes apoptosis by downregulating PRR11 expression and upregulating miR-144 in non-small cell lung cancer. Biomed Pharmacother. 2019;109:1851–9. https://doi.org/10.1016/j.biopha.2018.09.151.
Dong X, Chen R, Lin H, Lin T, Pan S. lncRNA BG981369 inhibits cell proliferation, migration, and invasion, and promotes cell apoptosis by SRY-related high-mobility group box 4 (SOX4) signaling pathway in human gastric Cancer. Med Sci Monit. 2018;24:718–26. https://doi.org/10.12659/MSM.905965.
R. C. Team. A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2013. http://www.r-project.org/.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(D1):D447–52. https://doi.org/10.1093/nar/gku1003.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integratedmodels of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. https://doi.org/10.1101/gr.1239303.
Torre LA, Siegel RL, Jemal A. Lung Cancer statistics. Adv Exp Med Biol. 2016;893:1–19. https://doi.org/10.1007/978-3-319-24223-1_1.
Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. https://doi.org/10.1016/S0140-6736(16)30958-8.
Lemjabbar-Alaoui H, Hassan OU, Yang YW, Buchanan P. Lung cancer: biology and treatment options. Biochim Biophys Acta. 2015;1856(2):189–210. https://doi.org/10.1016/j.bbcan.2015.08.002.
Denisov EV, Schegoleva AA, Gervas PA, Ponomaryova AA, Tashireva LA, Boyarko VV, et al. Premalignant lesions of squamous cell carcinoma of the lung: the molecular make-up and factors affecting their progression. Lung Cancer. 2019;135:21–8. https://doi.org/10.1016/j.lungcan.2019.07.001.
Prelaj A, Tay R, Ferrara R, Chaput N, Besse B, Califano R. Predictive biomarkers of response for immune checkpoint inhibitors in non-small-cell lung cancer. Eur J Cancer. 2019;106:144–59. https://doi.org/10.1016/j.ejca.2018.11.002.
Li Y, Han X, Feng H, Han J. Long noncoding RNA OIP5-AS1 in cancer. Clin Chim Acta. 2019;499:75–80. https://doi.org/10.1016/j.cca.2019.08.031.
Wang M, Zhou L, Yu F, Zhang Y, Li P, Wang K. The functional roles of exosomal long non-coding RNAs in cancer. Cell Mol Life Sci. 2019;76(11):2059–76. https://doi.org/10.1007/s00018-019-03018-3.
Huang Z, Lei W, Tan J, Hu HB. Long noncoding RNA LINC00961 inhibits cell proliferation and induces cell apoptosis in human non-small cell lung cancer. J Cell Biochem. 2018;119(11):9072–80. https://doi.org/10.1002/jcb.27166.
Xu Y, Li J, Wang P, Zhang Z, Wang X. LncRNA HULC promotes lung squamous cell carcinoma by regulating PTPRO via NF-κB. J Cell Biochem. 2019;120(12):19415–21. https://doi.org/10.1002/jcb.29119.
Wang B, Jiang H, Wang L, Chen X, Wu K, Zhang S, et al. Increased MIR31HG lncRNA expression increases gefitinib resistance in non-small cell lung cancer cell lines through the EGFR/PI3K/AKT signaling pathway. Oncol Lett. 2017;13(5):3494–500. https://doi.org/10.3892/ol.2017.5878.
Li J, Xia R, Liu T, Cai X, Geng G. LncRNA-ATB promotes lung squamous carcinoma cell proliferation, migration, and invasion by targeting microRNA-590-5p/NF90 Axis. DNA Cell Biol. 2020;39(3):459–73. https://doi.org/10.1089/dna.2019.5193.
Li J, Li H, Lv X, Yang Z, Gao M, et al. Polymorphism in lncRNA AC016683.6 and its interaction with smoking exposure on the susceptibility of lung cancer. Cancer Cell Int. 2018;4(18):91.
Lv X, Cui Z, Li H, Li J, Yang Z, Bi Y, et al. Polymorphism in lncRNA AC008392.1 and its interaction with smoking on the risk of lung cancer in a Chinese population. Cancer Manag Res. 2018;10:1377–87. https://doi.org/10.2147/CMAR.S160818.
Chen Y, Pan Y, Ji Y, Sheng L, Du X. Network analysis of differentially expressed smoking-associated mRNAs, lncRNAs and miRNAs reveals key regulators in smoking-associated lung cancer. Exp Ther Med. 2018;16(6):4991–5002. https://doi.org/10.3892/etm.2018.6891.
Wu DM, Wang YJ, Han XR, Wen X, Wang S, Shen M, et al. LncRNA LINC00880 promotes cell proliferation, migration, and invasion while inhibiting apoptosis by targeting CACNG5 through the MAPK signaling pathway in spinal cord ependymoma. J Cell Physiol. 2018;233(9):6689–704. https://doi.org/10.1002/jcp.26329.
Schlienger S, Ramirez RA, Claing A. ARF1 regulates adhesion of MDA-MB-231 invasive breast cancer cells through formation of focal adhesions. Cell Signal. 2015;27(3):403–15. https://doi.org/10.1016/j.cellsig.2014.11.032.
Acknowledgements
Special thanks to all participants who took part in this study.
Data availability statement
All data are fully available without restriction.
Funding
None.
Author information
Authors and Affiliations
Contributions
RJZ and MLH carried out the study design, analysis and interpretation of data. RJZ, MMW and MDZ drafted the manuscript. FJL contributed to collection of relevant literature and revision of the manuscript. All read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participant
The study was performed in accordance with the guidelines of the Declaration of Helsinki. The study protocol was approved by the Ethics Committee of the Institutional Review Board of the Cixi People’s Hospital and Ningbo Urology and Nephrology Hospital, Ningbo Yinzhou No 2. Hospital.
Consent for publication
Not applicable.
Competing interests
No competing interests exist.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1 Fig S1.
Three lncRNAs based risk score distribution, patients’ event-free survival time and a heatmap of the expression profiles of the three lncRNA. (a) Red dots represent the high-risk group and green dots represent the low-risk group (b) Red dots represent the dead group and green dots represent the alive group: a population-based study with a mean follow-up of 3.5 years.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zheng, R., Zheng, M., Wang, M. et al. Identification of a prognostic long noncoding RNA signature in lung squamous cell carcinoma: a population-based study with a mean follow-up of 3.5 years. Arch Public Health 79, 61 (2021). https://doi.org/10.1186/s13690-021-00588-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13690-021-00588-2