Abstract
Background
Quinolones are commonly used for treatment of infections by bacteria of the Enterobacteriaceae family. However, the rising resistance to quinolones worldwide poses a major clinical and public health risk. This study aimed to characterise a novel multiple resistance plasmid carrying three plasmid-mediated quinolone resistance genes in Escherichia coli clinical stain RJ749.
Methods
MICs of ceftriaxone, cefepime, ceftazidime, ciprofloxacin, and levofloxacin for RJ749 and transconjugant c749 were determined by the Etest method. Conjugation was performed using sodium azide-resistant E. coli J53 strain as a recipient. The quinolone resistance-determining regions of gyrA, gyrB, parC, and parE were PCR-amplified.
Results
RJ749 was highly resistant to quinolones, while c749 showed low-level resistance. S1-nuclease pulsed-field gel electrophoresis revealed that RJ749 and c749 both harboured a plasmid. PCR presented chromosomal mutation sites of the quinolone resistance-determining region, which mediated quinolone resistance. The c749 genome comprised a single plasmid, pRJ749, with a multiple resistance region, including three plasmid-mediated quinolone resistance (PMQR) genes (aac (6′)-Ib-cr, qnrS2, and oqxAB) and ten acquired resistance genes. One of the genes, qnrS2, was shown for the first time to be flanked by two IS26s. Three IS26-mediated circular molecules carrying the PMQR genes were detected.
Conclusions
We revealed the coexistence of three PMQR genes on a multiple resistance plasmid and a new surrounding genetic structure of qnrS2 flanked by IS26 elements. IS26 plays an important role in horizontal spread of quinolone resistance.
Similar content being viewed by others
Background
Quinolones are widely used for empirical and directed therapy in infections caused by members of the Enterobacteriaceae family [1]. The rise in global resistance to quinolones poses a great challenge to clinical treatment and public health, given the possible transmission of fluoroquinolone resistance from animals to humans [2]. In 2017, the resistance rate of Escherichia coli to ciprofloxacin was 57.0% in China [3].
Mechanisms of fluoroquinolone resistance occur principally through chromosomal mutations in the quinolone resistance-determining region (QRDR) of DNA gyrase (gyrA and gyrB) and DNA topoisomerase IV (parC and parE) and, to a lesser extent, changes in outer membrane proteins or efflux pumps [4, 5]. Moreover, plasmid-mediated quinolone resistance (PMQR) determinants have also been reported to encode different proteins, including Qnr proteins, the aminoglycoside acetyl transferase aac (6′)-Ib-cr, and efflux pumps qepA and oqxAB [6,7,8]. The emergence of PMQR genes has been reported, conferring low-level resistance to fluoroquinolones [2].
Qnr proteins bind to DNA gyrase and topoisomerase IV, protecting them from quinolone inhibition. The qnr gene, known as the ‘qnrA1’ allele, was the first plasmid-borne quinolone resistance gene to be described [9]. Subsequently, several classes of qnr genes (qnrB, qnrC, qnrD, qnrS, and qnrVC) that reduce susceptibility to fluoroquinolones have been identified [10, 11]. The aac (6′)-Ib-cr enzyme is a variant of aac (6′)-Ib. An acetylation assay showed the capacity of aac (6′)-Ib-cr to acetylate ciprofloxacin at the amino nitrogen on its piperazinyl substituent [6, 12,13,14]. It was selectively acting on ciprofloxacin and norfloxacin, which both have piperazinyl secondary amines. The qepA determinant was described in Escherichia coli clinical isolates from Japan and Belgium in 2007 and encodes an MFS-type efflux pump, which is able to excrete quinolone into the extracellular space [15]. The efflux pump oqxAB belongs to the resistance-nodulation-division (RND) family and is encoded by the oqxA and oqxB genes. It confers resistance to quinoxalines, chloramphenicol, trimethoprim, and quinolones [14]. These mechanisms cause low-level quinolone resistance but facilitate the emergence of higher-level resistance to quinolones at therapeutic levels.
In the current study, we discovered a novel plasmid with a multidrug resistance region (MRR), which comprised three kinds of plasmid-mediated quinolone resistance genes (qnrS2, aac (6′)-Ib-cr, and oqxAB).
Methods
Bacterial strains and antimicrobial susceptibility testing
Escherichia coli strain RJ749 was collected from Ruijin Hospital of Shanghai, China. The strain was isolated from a blood specimen. The MICs of ceftriaxone, cefepime, ceftazidime, ciprofloxacin, and levofloxacin were determined using Etest, and the results were interpreted based on the guidelines of the Clinical and Laboratory Standards Institutes [CLSI] (2018).
Conjugal transfer experiments, S1-nuclease pulsed-field gel electrophoresis (S1-PFGE), and plasmid typing
Conjugal transfer experiments were performed in broth culture using the strain RJ749 as the donor and the strain E. coli J53 (sodium azide-resistant) as the recipient. Selection was performed with 100 μg/mL concentrations of sodium azide and 2 μg/mL concentrations of cefotaxime. RJ749 and the transconjugant, c749, were both digested with S1 nuclease and subjected to S1-PFGE, as described previously [16].
PCR amplification, DNA sequencing, and sequence analysis
PCR assays were conducted targeting QRDRs of the gyrA, gyrB, parC, and parE genes with primer pairs as previously reported [17]. The genomic DNA was extracted using the QIAGEN Midi Kit (Qiagen, Hilden, Germany). The transconjugant c749 was sequenced using the Pacbio RS II (Pacific Biosciences, Menlo Park, CA, United States). The reads were de novo assembled using the HGAP 3.0 SMRTTM Pipe. Sequence similarity searches were carried out using BLAST [17]. Open reading frames (ORFs) were predicted using the ORF Finder software [18]. Protein-coding genes were initially identified and annotated using RAST. The antimicrobial resistance genes were identified using ResFinder [19]. Plasmid incompatibility groups were identified using PlasmidFinder and pMLST.
Identification of IS26-mediated formation of circular structures
Because IS26-flanked structures can generate circular DNA intermediates, reverse PCR was performed to detect potential circular forms. A series of PCR primers at sites flanking IS26 was used to determine the order of the circular structures carrying quinolone resistance determinants (Table 1).
Results
Early characterisation of RJ749 and c749
The MICs of cefotaxime, ceftriaxone, cefepime, ceftazidime, ciprofloxacin, and levofloxacin for RJ749 were ≥ 256, ≥ 256, ≥ 256, ≥ 32, and ≥ 32 mg/L, respectively. The MICs of cefotaxime, ceftriaxone, cefepime, ceftazidime, ciprofloxacin, and levofloxacin for c749 were ≥ 256, ≥ 256, 24, 12, 2, and 1 mg/L, respectively.
Transfer of multiple drug resistance
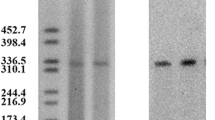
Through mating experiments, cephalosporin and quinolone resistance was transferred from the donor, RJ749, to the recipient, J53 (sodium azide-resistant). S1-PFGE revealed that RJ749 and c749 both harboured a plasmid, about 250 kbp (Fig. 1).
General genomic features
In the process of amplifying the QRDRs of the gyrA, gyrB, parC, and parE genes, chromosomal mutation sites of the quinolone resistance-determining region were determined, including gyrA and parC (83 and 87 sites of gyrA, 80 and 84 sites of parC), which mediated resistance to quinolone.
Whole genome sequencing was carried out for the transconjugant c749. The genome of c749 comprised a single plasmid, pRJ749, which was 253,002 bp in length and with a GC content of 47%, comprising 769 putative ORFs. pRJ749 had a multiple resistance region at positions 192,481–244,472, including for aminoglycosides (aadA16, aac (3)-IIa, aac (6′)-Ib-cr); β-lactams (blaOXA-1, blaTEM-1B); quinolones (aac (6′)-Ib-cr, qnrS2, oqxAB); macrolide-lincosamide-streptavidin B (MLS) (mph(A)); chloramphenicol (catB3); rifampicin class (ARR-3); sulfonamides (sul1); tetracyclines (tet(A)); and trimethoprim (dfrA27). Results of PlasmidFinder and pMLST revealed the IncHI2/−IncN multireplicon plasmid, in which IncHI2 belonged to ST3.
Sequence analysis indicated that pRJ749 belonged to the IncHI2-ST3 plasmid, in which three PMQR genes were located in a 51,991-bp multiple resistance region, and the MRR included ten additional resistance genes. Interestingly, blaCTX-M-64 is not located in the MRR. Instead, it inserted separately into the rest of this plasmid. It was reported that the CTX-M-64 enzyme is the result of recombination between members of the blaCTX-M-9 and blaCTX-M-1 derived genes, probably blaCTX-M-14 and blaCTX-M-15 [20].
Comparison of pRJ749 with other plasmids (Fig. 2) identified through BLAST revealed similar identities with p1 (84% coverage, 99% identity), pHS13-IncHI2 (90% coverage, 99% identity), and pMCR1_025943 (88% coverage, 99% identity). pHS13-IncHI2 and pMCR1_025943 belonged to ST3-IncHI2/−IncN, and p1 belonged to ST3-IncHI2.
Structures of circular forms mediated by IS26
Three circular forms carrying quinolone resistance genes and a single IS26 (IS26-intI1-aac (6′)-Ib-cr-blaOXA-1-catB-ARR3-qacEΔ1-Δsul1, IS26-qnrS2 and IS26-oqxAB) were amplified by reverse PCR and sequenced, respectively. These circular structures and sequences were identical to those of the corresponding region in plasmid pRJ749. Sequence analysis of the three circular molecules revealed the structures of a single IS26 and three PMQR genes, respectively (Fig. 3).
Region containing PMQR genes, including qnrS2, aac (6′)-Ib-cr, and oqxAB, is identical between pRJ749 and pHS13-IncHI2. The conserved core genes are indicated by grey arrows. Mobile elements and antimicrobial resistance genes are shown in red and yellow, respectively. Shaded area indicates 100% identity
Discussion
In this study, RJ749 was highly resistant to quinolones. PCR and plasmid sequencing analysis revealed the presence of chromosomal mutations and PMQR genes respectively. This is the first report of the coexistence of three PMQR genes on a multiple resistance plasmid. Studies showed that PMQR was usually low level, and its MIC value was below the CLSI resistance breakpoint [21]. The MIC of c749 demonstrated intermediate resistance, which indicated that the coexistence of three PMQR genes can cause an increase in MIC more than the presence of one or two PMQR genes reported [22, 23].
Based on BLAST, another E. coli strain HS13 (GenBank accession no. CP026492.1) isolated from Hong Kong, China, also carried the same three PMQR genes as we found (Fig. 3). This indicates that the plasmid carrying three PMQR genes is beginning to appear and spread in China. China is one of the countries with the highest fluoroquinolone resistance globally. Various PMQR genes have high prevalence in Enterobacteriaceae isolated from animals or clinically [24,25,26], challenging the use of fluoroquinolones, which needs to be taken seriously.
The oqxAB genes are often located on conjugative plasmids, within a composite transposon Tn6010 flanked by IS26 [20, 27,28,29,30,31]. In our study, the genetic environment of oqxAB was the same as that of the previously reported gene—flanked by two IS26s. In a previous study, we collected 125 E. coli clinical isolates and determined the genetic environment in six oqxAB-positive strains; the circular forms of IS26-oqxAB were detected in all six strains (data not shown). oqxAB was mainly reported in IncHI2-ST1/2 plasmids in Asia [31, 32]. Nevertheless, it has been reported that most IncHI2-oqxAB-blaCTX-M-9 plasmids in E. coli belong to IncHI2-ST3 [33]. oqxAB in our study was located on the IncHI2-ST3 plasmid in E. coli, which suggested that IncHI2 plasmids undergo constant evolvement and dissemination.
The aac (6′)-Ib-cr gene is usually found in a cassette as part of an integron in a multi-resistance plasmid, which may contain other PMQR genes [34, 35], and IS26 is often associated [36]. In pRJ749, aac (6′)-Ib-cr was located at a cluster of resistance genes flanked by IS26, which consisted of the aac (6′)-Ib-cr, blaOXA-1, catB3, arr-3, qacEΔ1, and Δsul1 elements as previously reported [37]. This cassette structure is widely present in Enterobacteriaceae in China, as demonstrated by BLAST.
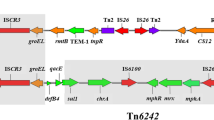
qnrS2 is mainly found on the IncQ [38] and IncU [39] plasmids of aquatic bacteria (mainly Aeromonas spp.). Two different surrounding genetic structures have been described for the qnrS2 genes. Two repC- and repA-like genes were located immediately downstream of the qnrS2 gene in two plasmids [39, 40]. Another genetic environment is part of a mobile insertion cassette (mic), the insertion of which disrupted an mpR gene encoding a putative zinc-metalloprotease (MpR) [41]. In 2016, a report in Shanghai, China, showed that the E. coli detection rate of qnrS2 in faeces of normal people was 3.7% [24]. Our study is the first to report qnrS2 flanked by two IS26s (Fig. 4). qnrS2 from E. coli strain HS13 (GenBank accession no. CP026492.1) isolated from Hong Kong, China, was also flanked by IS26s. This suggested that IS26 played an important role in the transfer of qnrS2 genes in plasmids among Enterobacteriaceae in China.
Three arrangements of surrounding genetic structures of qnrS2 gene. Open reading frames (ORFs) are shown as arrows, indicating the direction of transcription. Plasmid scaffolds, including the plasmid replicons, maintenance genes, and transfer region, are shown in violet, green, and blue, respectively. The gene qnrS2 is shown in yellow. Inverted repeats (IRL, left inverted repeat; IRR, right inverted repeat) are shown in boxes, with black; their length and sequence are shown below the structures. Direct repeats (DR) flanking the mobile insertion cassette are shown above the map. Mobile IS26 elements are shown in red. Homologous regions are indicated by grey shading
IS26 was observed to be predominant in the plasmids of Enterobacteriaceae [42]. Harmer and Hall [43] proposed a model in which the movement of IS26-associated antibiotic resistance genes occurred via a circular molecule that includes a single IS26 together with resistance genes. Recently, the insertion and excision of various IS26-derived circular molecules were reported in a single plasmid in a strain recovered from a chicken on a farm in Shangdong Province, China [44]. To investigate whether such circular molecules were present in the RJ749 strain, reverse PCR was performed with primers located within three PMQR genes and IS26s. Three amplicons were obtained, suggesting the presence of circular molecules. These findings strongly suggested that the IS26-associated PMQR genes can exist in circular and linear forms in the RJ749 strain. Harmer and Hall [45] reported the loss of IS26-associated antibiotic resistance mediated by plasmids R388::Tn4352B and R388::Tn4352 after five cycles of serial subculture. However, when we subcultured c749 in new broth for five cycles (data not shown), no significant change in quinolone susceptibility was found. Reverse PCR verified that PMQR genes remained in circular and linear forms, indicating that IS26-mediated quinolone resistance is stable.
Conclusions
In conclusion, a plasmid carrying three PMQR genes has been emerging in E. coli in China, and a new surrounding genetic structure of qnrS2 flanked by IS26 elements was identified for the first time. IS26 mediates the formation of circular intermediates, which results in various forms and horizontal spread of quinolone resistance.
Availability of data and materials
The plasmid sequence of pRJ749 obtained in this study has been deposited into GenBank with accession number MH491004.1. The accession numbers for the sequences used for comparison are as follows: p1, LT795115.1; pHS13-IncHI2, CP026492.1; pMCR1_025943, CP027202.2. The sequence of RJ749 has been deposited in the Sequence Read Archive (SRA) under the accession number SRR8195918.
Abbreviations
- MRR:
-
Multidrug resistance region
- ORFs:
-
Open reading frames
- PMQR:
-
Plasmid-mediated quinolone resistance
- QRDR:
-
Quinolone resistance-determining region
- S1-PFGE:
-
S1-nuclease pulsed-field gel electrophoresis
References
JG. Emergence and dissemination of quinolone-resistant Escherichia coli in the community. %J Antimicrobial agents and chemotherapy. 1999;11.
Poirel L, Cattoir V, Nordmann P. Plasmid-mediated quinolone resistance; interactions between human, animal, and environmental ecologies. Front Microbiol. 2012;3:24.
Hu F, Zhu D, Wang F, Wang M. Current status and trends of antibacterial resistance in China. Clin Infect Dis. 2018;67:S128–S34.
Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev. 2009;22:664–89.
Drlica KHH, Kerns R, et al. Quinolones:action and resistance updated. Curr Top Med Chem. 2009;9:981–98.
Jacoby GA, Strahilevitz J, Hooper DC. Plasmid-mediated quinolone resistance. Microbiol Spectrum. 2014;2(5):PLAS-0006-2013.
Robicsek ASJ, Jacoby GA, et al. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. NatMed. 2006;12:83–8.
Yamane KW. SuzukiS etal. Newplasmid-mediatedfluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother. 2007;51:3354–60.
Martinez-Martinez L PA, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet. 1998;351:797–9.
Rodriguez-Martinez JM, Machuca J, Cano ME, Calvo J, Martinez-Martinez L, Pascual A. Plasmid-mediated quinolone resistance: two decades on. Drug Resist Updat. 2016;29:13–29.
Hooper DC, Jacoby GA. Mechanisms of drug resistance: quinolone resistance. Ann N Y Acad Sci. 2015;1354:12–31.
Jiang Y, Zhou Z, Qian Y, Wei Z, Yu Y, Hu S, et al. Plasmid-mediated quinolone resistance determinants qnr and aac (6′)-Ib-cr in extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in China. J Antimicrob Chemother. 2008;61:1003–6.
Briales A, Rodriguez-Martinez JM, Velasco C, de Alba PD, Rodriguez-Bano J, Martinez-Martinez L, et al. Prevalence of plasmid-mediated quinolone resistance determinants qnr and aac (6′)-Ib-cr in Escherichia coli and Klebsiella pneumoniae producing extended-spectrum beta-lactamases in Spain. Int J Antimicrob Agents. 2012;39:431–4.
Hansen LH, Jensen LB, Sorensen HI, Sorensen SJ. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J Antimicrob Chemother. 2007;60:145–7.
Perichon B, Courvalin P, Galimand M. Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob Agents Chemother. 2007;51:2464–9.
Wong MH, Kan B, Chan EW, Yan M, Chen S. IncI1 plasmids carrying various blaCTX-M genes contribute to ceftriaxone resistance in Salmonella enterica Serovar Enteritidis in China. Antimicrob Agents Chemother. 2016;60:982–9.
Kim SY, Lee SK, Park MS, Na HT. Analysis of the Fluoroquinolone antibiotic resistance mechanism of Salmonella enterica isolates. J Microbiol Biotechnol. 2016;26:1605–12.
Bi D, Xu Z, Harrison EM, Tai C, Wei Y, He X, et al. ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res. 2012;40:D621–6.
Bjorkeng EK, Hjerde E, Pedersen T, Sundsfjord A, Hegstad K. ICESluvan, a 94-kilobase mosaic integrative conjugative element conferring interspecies transfer of VanB-type glycopeptide resistance, a novel bacitracin resistance locus, and a toxin-antitoxin stabilization system. J Bacteriol. 2013;195:5381–90.
Norman A, Hansen LH, She Q, Sorensen SJ. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid. 2008;60:59–74.
Wang J, Guo ZW, Zhi CP, Yang T, Zhao JJ, Chen XJ, et al. Impact of plasmid-borne oqxAB on the development of fluoroquinolone resistance and bacterial fitness in Escherichia coli. J Antimicrob Chemother. 2017;72:1293–302.
Machuca J, Briales A, Labrador G, Diaz-de-Alba P, Lopez-Rojas R, Docobo-Perez F, et al. Interplay between plasmid-mediated and chromosomal-mediated fluoroquinolone resistance and bacterial fitness in Escherichia coli. J Antimicrob Chemother. 2014;69:3203–15.
Machuca J, Ortiz M, Recacha E, Diaz-De-Alba P, Docobo-Perez F, Rodriguez-Martinez JM, et al. Impact of AAC (6′)-Ib-cr in combination with chromosomal-mediated mechanisms on clinical quinolone resistance in Escherichia coli. J Antimicrob Chemother. 2016;71:3066–71.
Ni Q, Tian Y, Zhang L, Jiang C, Dong D, Li Z, et al. Prevalence and quinolone resistance of fecal carriage of extended-spectrum beta-lactamase-producing Escherichia coli in 6 communities and 2 physical examination center populations in Shanghai, China. Diagn Microbiol Infect Dis. 2016;86:428–33.
Cao TT, Deng GH, Fang LX, Yang RS, Sun J, Liu YH, et al. Characterization of quinolone resistance in Salmonella enterica from farm animals in China. J Food Prot. 2017;80:1742–8.
Wen Y, Pu X, Zheng W, Hu G. High prevalence of plasmid-mediated quinolone resistance and IncQ plasmids carrying qnrS2 gene in Bacteria from Rivers near hospitals and aquaculture in China. PLoS One. 2016;11:e0159418.
Kim HB, Wang M, Park CH, Kim EC, Jacoby GA, Hooper DC. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:3582–4.
Zhao J, Chen Z, Chen S, Deng Y, Liu Y, Tian W, et al. Prevalence and dissemination of oqxAB in Escherichia coli isolates from animals, farmworkers, and the environment. Antimicrob Agents Chemother. 2010;54:4219–24.
Wong MH, Chen S. First detection of oqxAB in Salmonella spp. isolated from food. Antimicrob Agents Chemother. 2013;57:658–60.
Rodriguez-Martinez JM. Diaz de Alba P, Briales a, Machuca J, Lossa M, Fernandez-Cuenca F, et al. contribution of OqxAB efflux pumps to quinolone resistance in extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae. J Antimicrob Chemother. 2013;68:68–73.
Li L, Liao X, Yang Y, Sun J, Li L, Liu B, et al. Spread of oqxAB in Salmonella enterica serotype Typhimurium predominantly by IncHI2 plasmids. J Antimicrob Chemother. 2013;68:2263–8.
Campos J, Mourao J, Marcal S, Machado J, Novais C, Peixe L, et al. Clinical Salmonella Typhimurium ST34 with metal tolerance genes and an IncHI2 plasmid carrying oqxAB-aac (6′)-Ib-cr from Europe. J Antimicrob Chemother. 2016;71:843–5.
Fang L, Li X, Li L, Li S, Liao X, Sun J, et al. Co-spread of metal and antibiotic resistance within ST3-IncHI2 plasmids from E. coli isolates of food-producing animals. Sci Rep. 2016;6:25312.
Pitout JD, Wei Y, Church DL, Gregson DB. Surveillance for plasmid-mediated quinolone resistance determinants in Enterobacteriaceae within the Calgary health region, Canada: the emergence of aac (6′)-Ib-cr. J Antimicrob Chemother. 2008;61:999–1002.
Sabtcheva S, Kaku M, Saga T, Ishii Y, Kantardjiev T. High prevalence of the aac (6′)-Ib-cr gene and its dissemination among Enterobacteriaceae isolates by CTX-M-15 plasmids in Bulgaria. Antimicrob Agents Chemother. 2009;53:335–6.
Ruiz E, Rezusta A, Saenz Y, Rocha-Gracia R, Vinue L, Vindel A, et al. New genetic environments of aac (6′)-Ib-cr gene in a multiresistant Klebsiella oxytoca strain causing an outbreak in a pediatric intensive care unit. Diagn Microbiol Infect Dis. 2011;69:236–8.
Wong MH, Chan EW, Xie L, Li R, Chen S. IncHI2 plasmids are the key vectors responsible for oqxAB transmission among Salmonella species. Antimicrob Agents Chemother. 2016;60:6911–5.
Bonemann G, Stiens M, Puhler A, Schluter A. Mobilizable IncQ-related plasmid carrying a new quinolone resistance gene, qnrS2, isolated from the bacterial community of a wastewater treatment plant. Antimicrob Agents Chemother. 2006;50:3075–80.
Dobiasova H, Videnska P, Dolejska M. Complete sequences of IncU plasmids harboring quinolone resistance genes qnrS2 and aac (6′)-Ib-cr in Aeromonas spp. from ornamental fish. Antimicrob Agents Chemother. 2016;60:653–7.
Han JE, Kim JH, Choresca CH Jr, Shin SP, Jun JW, Chai JY, et al. First description of ColE-type plasmid in Aeromonas spp. carrying quinolone resistance (qnrS2) gene. Lett Appl Microbiol. 2012;55:290–4.
Picao RC, Poirel L, Demarta A, Silva CS, Corvaglia AR, Petrini O, et al. Plasmid-mediated quinolone resistance in Aeromonas allosaccharophila recovered from a Swiss lake. J Antimicrob Chemother. 2008;62:948–50.
SR. P, SM. K, N. F, SO. J. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin Microbiol Rev. 2018:31:e00088–17.
Harmer CJ, Moran RA, Hall RM. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. MBio. 2014;5:e01801–14.
He DD, Zhao SY, Wu H, Hu GZ, Zhao JF, Zong ZY, et al. Antimicrobial resistance-encoding plasmid clusters with heterogeneous MDR regions driven by IS26 in a single Escherichia coli isolate. J Antimicrob Chemother. 2019;74:1511–6.
Harmer CJ, Hall RM. IS26-mediated precise excision of the IS26-aphA1a Translocatable unit. MBio. 2015;6:e01866–15.
Acknowledgments
Not applicable.
Funding
This work was funded by National Key R&D Program of China [grant number 2018YFE0102400] and National Natural Science Foundation of China (NSFC) [grant number 81772245].
Author information
Authors and Affiliations
Contributions
All authors listed made a substantial, direct, and intellectual contribution to the work and approved the manuscript for publication.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the ethics committee of Ruijin Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China and the Review.
Board exempted the requirement for written informed consent because this is a retrospective study that only focused on bacteria and did not affect the patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tao, Y., Zhou, K., Xie, L. et al. Emerging coexistence of three PMQR genes on a multiple resistance plasmid with a new surrounding genetic structure of qnrS2 in E. coli in China. Antimicrob Resist Infect Control 9, 52 (2020). https://doi.org/10.1186/s13756-020-00711-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-020-00711-y