Abstract
Background
The emergence and spread of New Delhi metallo-β-lactamase-producing Enterobacteriaceae has been a serious challenge to manage in the clinic due to its rapid dissemination of multi-drug resistance worldwide. As one main type of carbapenemases, New Delhi metallo-β-lactamase (NDM)is able to confer resistance to almost all β-lactams, including carbapenems, in Enterobacteriaceae. Recently, New Delhi metallo-β-lactamase-5 attracted extensive attention because of increased resistance to carbapenems and widespread dissemination. However, the dissemination mechanism of blaNDM-5 gene remains unclear.
Methods
A total of 224 carbapenem-resistant Enterobacteriaceae isolates (CRE) were collected from different hospitals in Zhejiang province. NDM-5-positive isolates were identified and subjected to genotyping, susceptibility testing, and clinical data analysis. We established the genetic location of blaNDM-5 with southern blot hybridisation, and analysed plasmids containing blaNDM-5 with filter mating and DNA sequencing.
Results
Eleven New Delhi metallo-β-lactamase-5 (NDM-5)-producing strains were identified, including 9 Escherichia coli strains, 1 Klebsiella pneumoniae strain, and 1 Citrobacter freundii strain. No epidemiological links for E. coli isolates were identified by multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE). S1-PFGE and southern blot suggested that the blaNDM-5 gene was located on a 46-kb IncX3-type plasmid in all isolates. Nine of the 11 isolates (81.8%) tested could successfully transfer their carbapenem-resistant phenotype to E. coli strain C600. Moreover, sequence analysis further showed that this plasmid possessed high sequence similarity to most of previously reported blaNDM-5-habouring plasmids in China.
Conclusion
The present data in this study showed the IncX3 type plasmid played an important role in the dissemination of blaNDM-5 in Enterobacteriaceae. In addition, to the best of our knowledge, this report is the first to isolate both E. coli and C. freundii strains carrying blaNDM-5 from one single patient, which further indicated the possibility of blaNDM-5 transmission among diverse species. Close surveillance is urgently needed to monitor the further dissemination of NDM-5-producing isolates.
Similar content being viewed by others
Background
Enterobacteriaceae, such as E.coli, K. pneumoniae and C. freundii, are important pathogens that cause human infections. Carbapenem antibiotics are used in the treatment of infections caused by multi-drug resistant Enterobacteriaceae. However, the emergence of Carbapenem-resistant Enterobacteriaceae (CRE) has been a serious challenge to manage in the clinic because of the rapid worldwide dissemination of multi-drug resistance [1]. As one main type of carbapenemases, New Delhi metallo-β-lactamase (NDM)is able to confer resistance to almost all β-lactams, including carbapenems, in Enterobacteriaceae. Since the first report of blaNDM-1, 17 variants of NDM enzymes (NDM-1 to NDM-17) have been identified among Gram-negative bacteria worldwide (http://www.ncbi.nlm.nih.gov/pathogens/submit_beta_lactamase/). Among NDM carbapenemases, New Delhi metallo-β-lactamase-5, first identified in an E. coli strain in the UK in 2011, attracted extensive attention because of increased resistance to carbapenems and broad-spectrum cephalosporins [2]. In addition, blaNDM-5 was reported to be carried in different incompatibility typing plasmids to transfer [3], such as IncF, IncN and IncX3. These plasmids are able to facilitate the dissemination of blaNDM-5 among the members of Enterobacteriaceae through horizontal gene transfer. NDM-5-producing isolates have been identified worldwide, such as in America [4], Australia [5], China [6], Denmark [7] and India [8]. Furthermore, NDM-5-positive strains were not only isolated from clinical specimens but also from animals, such as dogs [9], cats [10] and cows [11]. Worryingly, blaNDM-5 has also been identified in environmental samples [hospital sewage water [12] and urban river [13]], indicating its presence in the community. However, the dissemination mechanism of blaNDM-5 gene remains unclear.
In this study, we screened NDM-5-producing Enterobacteriaceae to elucidate the dissemination mechanism. In addition, to the best of our knowledge, this report is the first to isolate E. coli and C. freundii strains carrying blaNDM-5 from the same patient.
Methods
Bacterial strains
From Jun. 2016 to Sep. 2017, 224 carbapenem-resistant Enterobacteriaceae isolates, as determined by the agar dilution method according to the Clinical and Laboratory Standards Institute guidelines [14], were obtained from four hospitals in different locations in Zhejiang, China. In a retrospective study, common carbapenemase genes (blaKPC, blaIMP, blaVIM, blaOXA-48, and blaNDM) were amplified, and the positive products were sequenced; eleven NDM-5 producing strains were identified for further study. The NDM-5 producing strains were preliminarily identified by the VITEK 2 system (Sysmex-bioMérieux, Marcy l’Etoile, France) and further confirmed by whole genome sequencing. The characteristics of the isolates and related clinical data are shown in Table 1.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using broth microdilution method [14]. The antibiotics tested in this study were amikacin, aztreonam, cefepime, ceftazidime, ciprofloxacin, gentamicin, imipenem, minocycline, colistin and tigecycline. The results were analysed according to the CLSI guidelines [14], except tigecycline and colistin, for which the European Committee on Antimicrobial Susceptibility Testing breakpoints were used (http://www.eucast.org/clinical_breakpoints). E. coli ATCC 25922 was used as a quality control strain.
Bacterial genotyping
Pulsed-field gel electrophoresis (PFGE) was performed to analyse the clonal relatedness of the NDM-5 producing E. coli isolates according to the previous study [15]. Briefly, the isolates were digested by XbaI endonuclease, which was carried out with a CHEF-Mapper XA PFGE system (Bio-Rad, USA) with a 5–35 s linear ramp for 22 h at 6 V/cm and 14 °C. The PFGE profiles were analyzed with BioNumerics software (Applied Maths, Sint-Martens-Latern, Belgium). The Salmonella enterica serotype Braenderup H9812 was used as the size marker.
MLST was also performed for molecular typing. Bacterial genomic DNA was extracted from these isolates. Seven housekeeping genes of E. coli (adk, fumC, gyrB, icd, mdh, purA and recA), and K. pneumoniae (gapa, infb, mdh, pgi, phoe, rpob) were amplified by PCR, and the products were sequenced to analyse the ST.
Southern blot analysis and conjugation experiments
To determine the plasmid location of the blaNDM-5 gene, genomic DNA digested with S1-nuclease (TaKaRa, Japan) was electrophoresed on a CHEF-mapper XA pulsed-field gel electrophoresis (PFGE) system (Bio-Rad, USA) for 18 h at 14 °C with run conditions of 6 V/cm and pulse times from 2.16 s to 63.8 s. The DNA fragments were transferred to a positive-charged nylon membrane (Millipore, USA) and then hybridized with a digoxigenin-labeled NDM-5-specific probe. An NBT/BCIP color detection kit (Roche, Germany) was then used to detect the fragments. The Salmonella enterica serotype Braenderup H9812 was used as the size marker.
A filter-mating experiment was performed between the blaNDM-5-positive isolates and rifampicin-resistant E. coli C600 as the recipient strain [15]. Transconjugants were selected on Mueller-Hinton agar plates containing 500 mg/L rifampicin and 100 mg/L ampicillin. PCR sequencing and antimicrobial susceptibility testing of the transconjugants were subsequently carried out to confirm whether the plasmid was successfully transferred to the recipient.
Plasmids analysis
Plasmid extraction and analysis was performed as previously described [15]. Briefly, the plasmid DNA of strains was extracted using a QIAamp DNA MiniKit (Qiagen, Valencia, CA, USA) following the manufacturer’s recommendations. The plasmids were sequenced on an Illumina-Hiseq™ 2000 (Illumina Inc., San Diego, U.S.A) platform with 2 × 100 bp paired-end reads. Sequence reads were assembled using CLC Genomics Workbench software package (CLC Bio 8.0). Gaps of a representative plasmid were closed by standard PCR and Sanger sequencing according to previous study [16]. The RAST (Rapid Annotation using Subsystems Technology) annotation website server (http://rast.nmpdr.org/rast.cgi) was then used to annotate the genomes of the plasmid. The circular map of the pEC463-NDM5 plasmid was generated using the CGview server [17]. A comparison of pEC463-NDM5 and three related plasmids was performed with EasyFig 2.2.2 [18]. The rested plasmid sequences were mapped to the representative plasmid sequence with CLC genomics workbench version 8.0.
Incompatibility typing of the blaNDM plasmid was performed by PCR-based replicon typing [19, 20] and was further identified with the help of PlasmidFinder-1.3 server (https://cge.cbs.dtu.dk/services/PlasmidFinder/).
In addition, plasmid stability was determined [3]. Briefly, the blaNDM-5-positive isolates were individually streaked out in the MH agar, incubated at 37 °C for 24 h, and then transferred to a fresh MH agar. After repeating this procedure for 12 days, 12 individual colonies were randomly selected. Subsequently, the blaNDM-5 gene was screened by PCR and sequenced.
Nucleotide sequence accession number
The complete sequence of the plasmid pEC463-NDM5 (accession number MG545911), is deposited at DDBJ/EMBL/GenBank.
Results and discussion
Isolate characteristics and antimicrobial susceptibility testing
Among the 224 CRE isolates, 137 isolates were KPC-2 carbapenemase producers, eleven isolates were NDM-5 carbapenemase producers, four isolates carried blaIMP-1 gene, two isolates carried blaVIM-1 gene and two isolates carried blaNDM-1 gene. In addition, 68 isolates exited other unknown mechanism of carbapenem-resistance.
In this study, eleven NDM-5-producing isolates were further identified, including nine E. coli, one K. pneumoniae and one C. freundii. These isolates were all recovered from hospitalized patients. These patients were aged between 16 and 85 years, with an average age of 55 years, had different severities of illness (Table 1), and all had previously received broad-spectrum antibiotics. Notably, with both E. coli (EC418) and C. freundii strains (CF418) were isolated from the feces of one patient from haematology department. This patient was found to be a carrier of blaNDM-5-positive strains. In contrast, the other patients from whom blaNDM-5-carrying strains were isolated from blood, pus, ascites, urine or sputum were symptomatic. In addition, these patients had no recent history of travel or hospitalization abroad.
The antimicrobial susceptibility testing results showed that the blaNDM-5-positive isolates were resistant to carbapenems, third-generation cephalosporins, and cefperazone/sulbactam. These isolates were also resistant to fluoroquinolones (81.8%), aztreonam (36.4%), amikacin (36.4%), nitrofurantoin (45.4%) and tigecycline (18.2%). All isolates were susceptible to colistin. E.coli EC122 and K. pneumoniae KP387 strains were both resistant to tigecycline, suggesting that increased resistance phenotypes of blaNDM-5-postive isolates are increasing in clinics. In addition, other β-lactamase genes, such as those encoding CTX-M-24, CTX-M-55, CMY-42, were also frequently detected in various blaNDM-5-positive E. coli strains (Fig. 1). Gene encoding SHV-1 and CMY-26 were detected in the K. pneumoniae KP387 and C. freundii CF418 strains, respectively.
S1-digested plasmid DNA and southern blot hybridization of blaNDM-5 positive isolates. Bands in A with arrows pointing to them showed positive signals in Southern blot hybridization with the NDM-5 probe. M = Salmonella serotype Braenderup strain H9812 molecular marker. 1 = K. pneumoniae KP387; 2 = E. coli EC135; 3 = E. coli EC463; 4 = E. coli EC734; 5 = E. coli EC144; 6 = E. coli EC122; 7 = E. coli EC418; 8 = C. freundii CF418; 9 = E. coli EC310; 10 = E. coli EC611; 11 = E. coli EC126
Our recent studies showed that blaNDM-5 was able to coexist in the same isolate with tigecycline and colistin resistance phenotypes, thereby generating strains that approached pan-resistance. For example, blaNDM-5 was not only identified in high-level tigecycline resistance E. coli strains [21], but also coexisted in the same strain with the transferrable colistin resistance gene mcr-1 [15]. It is clear that generating strains results in so-called “superbug” isolates and accelerating entery into a “postantibiotic” era [22].
Genetic relatedness
MLST and PFGE experiments were performed to analyse the clonal relatedness of blaNDM-5-positive isolates because NDM-5 producers are infrequently isolated worldwide. According to the MLST results, nine blaNDM-5-postive E. coli isolates were grouped into 9 different sequence types. In accordance with the MLST results (Fig. 1), the different PFGE patterns confirmed that the seven E. coli isolates are not clonally related to each other even though some of the strains were collected from the same hospital. Strains EC122 and EC144 own similar the PFGE profiles, but the two strains have different sequence type and different resistance genes. Furthermore, core genome multi-locus sequence typing (cg-MLST) analysis in our study showed the blaNDM-5-positive isolates were not clonal relatedness (Additional file 1: Figure S1). In addition, the K. pneumoniae KP487 isolate belongs to ST182.
A previous study collected 11 NDM-5-producing E. coli strains from 7 hospitals in various locations in China from 2013 to 2014, and found that ST167 E. coli strains in clinical settings exhibited close linkages with the blaNDM-5 gene [23]. Our previous study also showed that high-level tigecycline resistance E. coli strains carrying blaNDM-5 also belonged to the ST167 clonal lineage [21], indicating that the ST167 sequence type is an important reservoir of blaNDM-5 in China. However, the diversity of MLST and PFGE types in the present study showed that the blaNDM-5 gene has been carried in other STs E. coli isolates from 2016 to 2017. Moreover, the blaNDM-5 gene was detected in the K. pneumoniae and one C. freundii strains, indicating that this gene has further disseminated in Enterobacteriaceae. Note that NDM-5-related outbreak has been reported [24, 25]. Although no genetic association was found between our blaNDM-5-positive isolates with other strains, the widespread dissemination of blaNDM-5 in recent years in Enterobacteriaceae highlights the need for extensive attention.
Location of the bla NDM-5 gene
S1-PFGE followed by Southern blot demonstrated that the blaNDM-5-positive strains were all located on plasmids of the same size(~ 46 Kb) (Fig. 2). The filter mating experiments were carried out to confirm the transferability of these blaNDM-5 plasmids. Nine of the 11 isolates tested could successfully transfer their carbapenem-resistant phenotype to E. coli strain C600 (Table 2). In addition,incompatibility plasmid classification showed that all the blaNDM-5 plasmids belonged to the IncX3-type plasmid. IncX3 plasmids might have played an important role in mediating the horizontal transmission of the blaNDM gene. This possibility has been supported by the results of several studies [6, 26–29]. In this study, blaNDM-5 was carried by the IncX3 plasmids. Moreover, 81.8% (9/11) of isolates carrying this type plasmid were able to transfer carbapenem-resistant phenotype. However, conjugation experiments of E. coli EC126 and EC135 strains were not performed because these two strains were resistant to rifampin. To date, IncX3 plasmids carrying blaNDM-5 have been reported worldwide [3, 22, 23]. Therefore, our present study further supplements those previous studies. In addition, we isolated E. coli and C. freundii strains carrying blaNDM-5 from a single patient. These blaNDM-5-carrying plasmids had very similar sequences (99% coverage and 98% similarity), indicating probable horizontal transfer of blaNDM-5 between E. coli and C. freundii strains by one same plasmid. In addition, the plasmid stability experiments showed that the blaNDM-5-positive plasmids were all stable in these isolates. After 12 rounds of subculture in MH agar without antibiotic addition, the randomly selected strains all carried the blaNDM-5 gene and a plasmid identical to their parental isolate in size. Overall, it is important for the IncX3 type plasmid to play an important role in the further dissemination of blaNDM-5 in Enterobacteriaceae. Therefore, it is imperative that effective measures be taken immediately to control the spread of this plasmid.
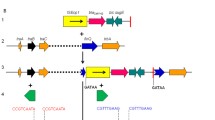
Plasmid sequence analysis of bla NDM-5
The entire plasmid sequence was obtained to better characterize the blaNDM-5-positive plasmid. Sequence analysis showed that the plasmid was 46,145 bp in length (Fig. 3a). The blaNDM-5 gene was preceded by IS3000, ISAba125 and IS5, and followed by bleMBL, trpF, dsbC, IS6 and ISkox3.No other antimicrobial resistance genes were detected in this plasmid.
Plasmid analysis of pEC463-NDM5. Schematic map of plasmid p pEC463-NDM5 (a), comparative analysis of three blaNDM-5-carrying IncX3 plasmids (b). The putative open reading frames are shown as arrowheads orrods (less than 130 amino acids). The gene name is shown near the corresponding arrowhead or rod. The depthof shading is indicative of the percentage BLASTN match, as indicated on the bottom
Further sequence alignments based on BLAST revealed that the plasmid sequences showed almost identical nucleotide sequences with those of the previously reported IncX3 plasmids pNDM-MGR194 of K. pneumoniae MGR-K194 in India [8]. The plasmid pNDM-MGR194 carrying blaNDM-5 was reported in 2015 in India, which was considered to play an important role in the dissemination of the blaNDM-5 gene because pNDM-MGR194-like plasmid was highly similar to those plasmids reported in China [3], Australia [5] and Denmark [7]. In addition, most of the blaNDM-5-carrying plasmids reported in China belonged to the IncX3-type and were identical or near-identical to pNDM-MGR194-like plasmid (Table 3). In this study, identification of the IncX3-type pNDM-MGR194-like plasmid in E. coli of different STs, K. pneumoniae and C. freundii strains indicated that this plasmid could mediate inter- and intra-species transfer of blaNDM-5. This possibility was further supported by our conjunction experimental data in vitro. Moreover, this plasmid carried in E. coli and C. freundii strains was isolated from faeces sample of a single patient at the same time, providing strong evidence that this plasmid could mediate blaNDM-5 dissemination in Enterobacteriaceae. Overall, our results revealed that IncX3-type pNDM-MGR194-like plasmids facilitate the rapid dissemination of blaNDM-5 among Enterobacteriaceae in China.
Conclusions
We report a near-term epidemiological study demonstrating the further dissemination of Enterobacteriaceae with the blaNDM-5 gene in China. Our work provides evidence that the IncX3-type plasmid played an important role in the dissemination of blaNDM-5 in Enterobacteriaceae. In addition, to the best of our knowledge, this report is the first to isolate E. coli and C. freundii strains carrying blaNDM-5 from a single patient. Close surveillance is urgently needed to monitor the further spread of NDM-5-producing isolates.
Abbreviations
- cg-MLST:
-
Core genome multi-locus sequence typing
- CLSI:
-
Clinical & Laboratory Standards Institute
- CRE:
-
Carbapenem-resistant Enterobacteriaceae isolates
- MIC:
-
Minimum inhibitory concentration
- MLST:
-
Multilocus sequence typing
- NDM:
-
New Delhi metallo-β-lactamase
- PFGE:
-
Pulsed-field gel electrophoresis
- RAST:
-
Rapid Annotation using Subsystems Technology
References
Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev. 2012;25(4):682–707.
Hornsey M, Phee L, Wareham DW. A novel variant, NDM-5, of the New Delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother. 2011;55(12):5952–4.
Zhu YQ, Zhao JY, Xu C, Zhao H, Jia N, Li YN. Identification of an NDM-5-producing Escherichia coli sequence type 167 in a neonatal patient in China. Sci Rep. 2016;6:29934.
de Man TJ, Perry KA, Avillan JJ, Rasheed JK, Limbago BM. Draft genome sequence of a New Delhi Metallo-beta-Lactamase-5 (NDM-5)-producing multidrug-resistant Escherichia coli isolate. Genome Announc. 2015;3(2):e00017–15.
Wailan AM, Paterson DL, Caffery M, Sowden D, Sidjabat HE. Draft genome sequence of NDM-5-producing Escherichia coli sequence type 648 and genetic context of blaNDM-5 in Australia. Genome Announc. 2015;3(2):e00194–15.
Yang P, Xie Y, Feng P, Zong Z. blaNDM-5 carried by an IncX3 plasmid in Escherichia coli sequence type 167. Antimicrob Agents Chemother. 2014;58(12):7548–52.
Hammerum AM, Littauer P, Hansen F. Detection of Klebsiella pneumoniae co-producing NDM-7 and OXA-181, Escherichia coli producing NDM-5 and Acinetobacter baumannii producing OXA-23 in a single patient. Int J Antimicrob Agents. 2015;46(5):597–8.
Hammerum M, Kamatchi C, Jha AK, Devasena N, Vennila R, Sumathi G, Vaidyanathan R. Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian J Med Microbiol. 2015;33(1):30–8.
Yousfi M, Mairi A, Bakour S, Touati A, Hassissen L, Hadjadj L, Rolain JM. First report of NDM-5-producing Escherichia coli ST1284 isolated from dog in Bejaia, Algeria. New Microbes New Infections. 2015;8:17–8.
Yousfi M, Touati A, Mairi A, Brasme L, Gharout-Sait A, Guillard T, De Champs C. Emergence of Carbapenemase-producing Escherichia coli isolated from companion animals in Algeria. Microbial Drug Resistance (Larchmont, NY). 2016;22(4):342–6.
He T, Wei R, Zhang L, Sun L, Pang M, Wang R, Wang Y. Characterization of NDM-5-positive extensively resistant Escherichia coli isolates from dairy cows. Vet Microbiol. 2017;207:153–8.
Parvez S, Khan AU. Hospital sewage water - a reservoir for variants of New Delhi metallo-beta-lactamase (blaNDM) and ESBL-producing enterobacteriaceae. Int J Antimicrob Agents. 2018;51(1):82–88.
Almakki A, Maure A, Pantel A, Romano-Bertrand S, Masnou A, Marchandin H, Jumas-Bilak E, Licznar-Fajardo P. NDM-5-producing Escherichia coli in an urban river in Montpellier, France. Int J Antimicrob Agents. 2017;50(1):123–4.
CLSI, editor. Performance standards for antimicrobial susceptibility testing; 27th ed. CLSI supplement M100. Wayne: Clinical and Laboratory Standards Institute; 2017.
Quan J, Li X, Chen Y, Jiang Y, Zhou Z, Zhang H, Sun L, Ruan Z, Feng Y, Akova M, et al. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017;17(4):400–10.
Fu Y, Liu L, Li X, Chen Y, Jiang Y, Wang Y, Yu Y, Xie X. Spread of a common blaNDM-1-carrying plasmid among diverse Acinetobacter species. Infect Genet Evol. 2015;32:30–3.
Grant JR, Stothard P. The CGView server. A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36(Web Server issue):W181–4.
Sullivan MJ, Petty NK, Beatson SA. Easyfig. a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–10.
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–28.
Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid. 2012;68(1):43–50.
Li X, Mu X, Yang Y, Hua X, Yang Q, Wang N, Du X, Ruan Z, Shen X, Yu Y. Rapid emergence of high-level tigecycline resistance in Escherichia coli strains harbouring blaNDM-5 in vivo. Int J Antimicrob Agents. 2016;47(4):324–7.
Bulman ZP, Chen L, Walsh TJ, Satlin MJ, Qian Y, Bulitta JB, Peloquin CA, Holden PN, Nation RL, Li J et al. Polymyxin Combinations Combat Escherichia coli Harboring mcr-1 and blaNDM-5: Preparation for a Postantibiotic Era. mBio. 2017;8(4):e00540–17.
Huang Y, Yu X, Xie M, Wang X, Liao K, Xue W, Chan EW, Zhang R, Chen S. Widespread dissemination of Carbapenem-resistant Escherichia coli sequence type 167 strains harboring blaNDM-5 in clinical settings in China. Antimicrob Agents Chemother. 2016;60(7):4364–8.
Hammerum AM, Hansen F, Olesen B, Struve C, Holzknecht BJ, Andersen PS, Thye AM, Jakobsen L, Roder BL, Stegger M, et al. Investigation of a possible outbreak of NDM-5-producing ST16 Klebsiella pneumoniae among patients in Denmark with no history of recent travel using whole-genome sequencing. J Global Antimicrob Resist. 2015;3(3):219–21.
Bathoorn E, Rossen JW, Lokate M, Friedrich AW, Hammerum AM. Isolation of an NDM-5-producing ST16 Klebsiella pneumoniae from a Dutch patient without travel history abroad, August 2015. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2015;20(41) https://doi.org/10.2807/1560-7917.ES.2015.20.41.30040.
Ho PL, Li Z, Lo WU, Cheung YY, Lin CH, Sham PC, Cheng VC, Ng TK, Que TL, Chow KH. Identification and characterization of a novel incompatibility group X3 plasmid carrying Bla NDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerging Microbes Infections. 2012;1(11):e39.
Sonnevend A, Al Baloushi A, Ghazawi A, Hashmey R, Girgis S, Hamadeh MB, Al Haj M, Pal T. Emergence and spread of NDM-1 producer Enterobacteriaceae with contribution of IncX3 plasmids in the United Arab Emirates. J Med Microbiol. 2013;62(Pt 7):1044–50.
Gottig S, Hamprecht AG, Christ S, Kempf VA, Wichelhaus TA. Detection of NDM-7 in Germany, a new variant of the New Delhi metallo-beta-lactamase with increased carbapenemase activity. J Antimicrob Chemother. 2013;68(8):1737–40.
Yang Q, Fang L, Fu Y, Du X, Shen Y, Yu Y. Dissemination of NDM-1-producing Enterobacteriaceae mediated by the IncX3-type plasmid. PLoS One. 2015;10(6):e0129454.
Chen D, Gong L, Walsh TR, Lan R, Wang T, Zhang J, Mai W, Ni N, Lu J, Xu J, et al. Infection by and dissemination of NDM-5-producing Escherichia coli in China. J Antimicrob Chemother. 2016;71(2):563–5.
Zheng B, Lv T, Xu H, Yu X, Chen Y, Li J, Huang C, Guo L, Zhang J, Jiang X et al. Discovery and characterization of an escherichia coli ST206 strain producing NDM-5 and MCR-1 from a patient with acute diarrhea. Int J Antimicrob Agents. 2018;51(2):273–5.
Zheng B, Xu H, Yu X, Lv T, Jiang X, Cheng H, Zhang J, Chen Y, Huang C, Xiao Y. Identification and genomic characterization of a KPC-2-, NDM-1- and NDM-5-producing Klebsiella michiganensis isolate. J Antimicrob Chemother. 2018;73(2):536–8.
Rahman M, Shukla SK, Prasad KN, Ovejero CM, Pati BK, Tripathi A, Singh A, Srivastava AK, Gonzalez-Zorn B. Prevalence and molecular characterisation of New Delhi metallo-beta-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. Int J Antimicrob Agents. 2014;44(1):30–7.
Li X, Jiang Y, Wu K, Zhou Y, Liu R, Cao Y, Wu A, Qiu Y. Whole-genome sequencing identification of a multidrug-resistant Salmonella enterica serovar typhimurium strain carrying blaNDM-5 from Guangdong, China. Infect Genet Evol. 2017;55:195–8.
Baraniak A, Izdebski R, Fiett J, Gawryszewska I, Bojarska K, Herda M, Literacka E, Zabicka D, Tomczak H, Pewinska N, et al. NDM-producing Enterobacteriaceae in Poland, 2012-14: inter-regional outbreak of Klebsiella pneumoniae ST11 and sporadic cases. J Antimicrob Chemother. 2016;71(1):85–91.
Pitart C, Sole M, Roca I, Roman A, Moreno A, Vila J, Marco F. Molecular characterization of blaNDM-5 carried on an IncFII plasmid in an Escherichia coli isolate from a nontraveler patient in Spain. Antimicrob Agents Chemother. 2015;59(1):659–62.
Nakano R, Nakano A, Hikosaka K, Kawakami S, Matsunaga N, Asahara M, Ishigaki S, Furukawa T, Suzuki M, Shibayama K, et al. First report of metallo-beta-lactamase NDM-5-producing Escherichia coli in Japan. Antimicrob Agents Chemother. 2014;58(12):7611–2.
Balm MN, La MV, Krishnan P, Jureen R, Lin RT, Teo JW. Emergence of Klebsiella pneumoniae co-producing NDM-type and OXA-181 carbapenemases. Clin Microbiol Infect. 2013;19(9):E421–3.
Acknowledgments
We would like to thank Long Sun (Hangzhou Hospital of Zhejiang Provincial Corps), Lihua Zhou (The First People’s Hospital of Huzhou) and Qing Lv (Shaoxing Hospital) for collecting partial isolates.
Funding
This study was supported by National Natural Science Foundation of China (31700125 and 81471986) and the Natural Science Young Foundation of Zhejiang Province, China (LQ17H190006) and the Medical and Health Research Project of Zhejiang Province, China (2017KY224). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
Please contact corresponding author for data requests.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: YY and DW; Performed the experiments: XL, YF and MS; Analyzed the data: DH XD, YZ and QH; Wrote the manuscript: XL and YF; All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not required.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
cg-MLST of blaNDM-5-positive isolates. (DOCX 61 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, X., Fu, Y., Shen, M. et al. Dissemination of blaNDM-5 gene via an IncX3-type plasmid among non-clonal Escherichia coli in China. Antimicrob Resist Infect Control 7, 59 (2018). https://doi.org/10.1186/s13756-018-0349-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-018-0349-6