Abstract
To develop a new antiviral preparation from a microbial source, the halophilic bacterium Bacillus pumilus E303035 was isolated from a soil sample collected at Qarhan Salt Lake in Qinghai, China. The inhibitory activity of an ethyl acetate extract of its fermentation broth was higher than that of an n-butanol extract. After isolation and purification, 9 compounds were obtained: cyclo(L-Leu-L-Pro) (1), cyclo(L-Pro-L-Tyr) (2), Brevianamide F (3), 2-(3-Indolyl) ethanol (4), N-[2-(1H-indol-3-yl) ethyl] acetamide (5), 3, 3-di(1H-indol-3-yl)propane-1,2-diol (6), Lincomycin B (7), dibutylphthalate (8), and p-hydroxyphenethyl alcohol (9). Compounds 1, 5, and 9 showed inhibitory activities against potato virus Y (PVY). Compounds 1, 4, and 9 had significant inhibitory activity against genes HC-pro, P3, and Nib, compound 5 against gene P3, and compounds 1 and 4 against NIa. Compounds 1, 4, 5, and 9 had significant inhibitory activity against genes VPg and 6K1. Active compounds 1, 5, and 9 had various effects on the expression of viral genes related to pathogenesis. Expression of genes cullin and XTH was up-regulated and CP was down-regulated, compared to the positive control. In conclusion, compounds 1, 5, and 9 might be considered as potential antiviral agents for future development.
Similar content being viewed by others
Introduction
The species Potato virus Y (PVY) is the most important viral pathogen affecting potato crops and a serious threat to potato production worldwide (Davie et al. 2017; Funke et al. 2017). It can infect various crops, including potato, tobacco, tomato, pepper and other crops belonging to the Solanaceae family, and can cause serious damage and significant economic loss (Zhang et al. 2017). Currently, few agents are available to effectively control this plant virus, such as ningnanmycin, dufulin, and ribavirin (Xiao et al. 2015). The search for new antiviral compounds is critical, by considering new sources and novel antiviral mechanisms.
Microbes are a broad and rich resource of natural bioactive products. Bacteria of the genus Bacillus are known to produce a wide range of active substances. They have successfully been used to generate compounds such as amylase, proteases, antibiotics, and surfactants (Caetano et al. 2011). The antagonistic effects of Bacillus are due to secondary metabolite production of antibiotics and antimicrobial peptides (Hu et al. 2010).
In this study, a halophilic Bacillus strain Bacillus pumilus E303035 with antiviral activity against PVY which we have screened in our lab was chosen. 9 compounds were isolated from this strain. The antiviral activities against PVY of fermentation extracts and isolated compounds were evaluated with real-time PCR. In order to illustrate the inhibitory mechanisms of active compounds isolated from strain E303035, the effects were determined on total RNA and on genes encoding the functional proteins of PVY.
Materials and methods
Plant and PVY materials
Tobacco plants (Nicotiana tabacum var. Samsun NN) were grown under greenhouse conditions, with 16 h light at 25 °C and 8 h darkness at 15 °C, and PVY was kept at 4 °C, in the Key Laboratory of Qinghai-Tibet Plateau Biotechnology (KLB), Ministry of Education, Qinghai Province, China. Three or four fully expanded leaves from, approximately, one-month-old plants were used in the experiment.
Strains and preparation of fermentation broth
Halophilic bacterium Bacillus pumilus Strain E303035 (Genbank accession number: MN238693; GDMCC number:60077) was isolated from a Qarhan Salt Lake soil sample, and was preserved at 4 °C in the KLB. It was cultured at 37 °C, with a rotation speed of 200 r/min in a shaker cabinet for 7 days, in Erlenmeyer flasks (1 L). Each flask contained 600 mL of ATCC213 medium (10 g MgSO4·7H2O, 0.2 g CaCl2·2H2O 0.2 g, KCl, 2.5 g peptone, 10 g yeast extract, 30 g NaCl, adding deuterium-depleted water (D.D.W) to 1000 mL, pH 7.2–7.4).
Then, the fermentation broth was centrifuged at a rotation speed of 8000 r/min, the supernatant was filtered through a 0.22 µm Millipore filter and stored at 4 °C for the preparation of crude extracts (Zhang et al. 2017).
Preparation of crude extracts of fermentation broth of strain E303035
The fermentation broth of strain E303035 was extracted with either ethyl acetate or n-butanol. The two kinds of extract were concentrated in vacuo to yield a residue. The residues were dissolved in distilled water (using DMSO as a vehicle, at a maximum concentration of 0.1%) to prepare concentrations of 1, 5, 10, and 20 mg/ml for antiviral testing against PVY (Zhang et al. 2017).
Isolation and identification of compounds in strain E303035
1H, 13C, DEPT, 1H-1H-COSY, HSQC, and HMBC NMR spectra were obtained with a JNM-ECA600 spectrometer. FAB-MS spectra were recorded on a JEOL JMS-HX 110 instrument. The chromatographic stationary phases were silica gel (200–300 mesh), Sephadex LH-20 (25–100 µm, Pharmacia) and MCI-gel CHP20P (75–150 mm, Mitsubishi Chemical). Using thin-layer chromatography, compounds were visualized by spraying with 5% H2SO4, followed by heating (Shen et al. 2013).
The ethyl acetate extract was dissolved in ethanol and concentrated in vacuo to yield 11 g oily fraction. Afterwards, the fraction was subjected to handera-SI (10 µm), and eluted with n-hexane: isopropanol in the range 80:20−50:50, to give Fraction 1 (Fr1) 4.5 g, Fr2 248 mg, Fr3 516 mg, Fr4 597 mg, and Fr5 267 mg.
Fr1 was subjected to MCI-gel CHP20P and eluted with 100% MeOH, MeOH:H2O (1:2), and 100% acetone to give compound 8 (3900 mg). Fr2 was subjected to Handera C18 (10 µm) and eluted with C2H3N-H2O (1:10–3:10) to give compounds 4 (51 mg) and 9 (10 mg). Fr3 was subjected to Handera C18 (10 µm) and eluted with C2H3N:H2O (1:20–7:20) to give compound 7 (13 mg). Fr4 was subjected to Handera C18 (10 µm) and eluted with C2H3N:H2O (1:20–3:10) to give compounds 1 (86 mg), 5 (78 mg), and 6 (8 mg). Fr5 was subjected to Handera C18 (10 µm) and eluted with C2H3N:H2O (1:20–3:10) to give compounds 2 (25 mg) and 3 (11 mg).
Compounds 1–9 were dissolved in distilled water (using DMSO as a vehicle, at a maximum concentration of 0.1%) to prepare the 31, 63, 125, 250, and 500 mg/ml concentrations for antiviral testing against PVY (Zhang et al. 2017).
PVY inoculation
Inoculation with PVY, and with PVY with an equal volume of extract (crude extracts, compounds, or ningnanmycin), was conducted on two top leaves of the tobacco plants, using a conventional friction method with a 10× concentration of inoculums. Phosphate buffer inoculation was used as a control. The two infected leaves were collected from each plant, for RNA extraction, 7 days from when the symptoms appeared (Chen et al. 2017).
Real-time PCR (qPCR) analysis
The total RNA was extracted using a Total RNA Reagent Kit (Takara) from the leaves of plants that were subjected to PVY infection after treatment with each extract (at concentrations of 1, 5, 10, 20 mg/mL, using DMSO as vehicle at a maximum concentration of 0.1%) or compound (at concentrations of 31, 63, 125, 250, and 500 μg/mL, using DMSO as a vehicle at maximum concentration of 0.1%), in triplicate. Total RNA samples were reverse transcribed using a PrimeScriptTMRT reagent Kit with gDNA Eraser (Takara). Real-time PCR was performed with SYBR Premix Ex Taq Kit (Takara), with β-actin as an internal control. The final volume of the PCR mixture was 20 µL, including 10 µL of the SYBR Green Master mix reagent, 7.8 µL of sterile water, 0.4 µL of DyeII, 1 µL of cDNA, and 0.4 µL (2 mM) of each real-time PCR primer. Primers used were Rβ-actin (5´-AAGGGATGCGAGGATGGA-3´) and Fβ-actin (5´-CAAGGAAATCACCGCTTTGG-3´), RPVY (5´-TTCATCTCCATCCATCATAACC-3´) and FPVY (5´-TACAACTTGCATACGACATAGG-3´), RLhc (5´-TTAAGAGAAGAAGCCGAATGTG-3´)and FLhc (5´-CCACACTTCAACTTGCTGAG-3´), RPSii (5´- TTTCTCCTCCCTCCCTTTCTCT-3´) and FPSii (5´-TTGCTTGACCGTCGTTGTG-3´), RXTH (5´-GCGAGGATTTGAGGCACAG-3´) and FXTH (5´-GCAACGAGAGGTGGATTAGAGAA-3´), RCPIP3 (5´-GTCAGTACAGCCAGAGCCAGAA-3´) and FCPIP3 (5´-AAGCGACTACCAAAACACACACA-3´), Rcullin-1 (5´-GTCGCAGAATGTGGCAAGAA-3´)and Fcullin-1 (5´-AGAGAAGAGAGATGTGGTTGGT TTG-3´). The following conditions were used: 95 °C for 10 s, 95 °C for 50 s, and 35 cycles of both 95 °C for 6 s and 62 °C for 35 s. Standard curves were constructed using a series of five tenfold dilutions of the cDNA template. The relative expression levels were calculated (Shen et al. 2018).
PCR analysis
The total RNA was extracted using a Total RNA Reagent Kit (Takara) from the leaves of plants that were subjected to PVY infection after the different treatments of compounds (at the concentrations of 31, 63, 125, 250, and 500 μg/mL, in triplicate. Total RNA samples were reverse transcribed using a PrimeScriptTMRT reagent Kit with gDNA Eraser (Takara). β-actin was used as an internal control. The final volume of the PCR mixture was 20 µL, including 10 µL of 2× FastTaq PCR Master Mix, 1µL of cDNA, 8.6 µL of sterile water, and 0.2µL (2 mM) of each RT-PCR primer. Primers used were Rβ-actin (5´-AAGGGATGCGAGGATGGA-3´) and Fβ-actin (5´-CAAGGAAATCACCGCTTTGG-3´), RHC-Pro (5´-ACCAACTCTATAGTGCTTAATGTCAGA-3´)and FHC-Pro (5´-GGAGTTCTAGACTCAATGGTTCAGT-3´), RP1 (5´-TTGTGTAACCTTGGAACGCGC-3´) and F P1 (5´-ATGGCAACTTACATGTCAACGATTC-3´), RP3 (5´-CTGATGCCGCACATTATATTCTTC-3´) and FP3 (5´-GGTATTCCTGGGCATGTCCTG-3´), RNIa (5´-TTGCTCTACAACAACATCATGATCAA-3´) and FNIa (5´-GCCAAATCACTCATGAGAGGTTTAA-3´), RNIb (5´-TTGATGGTGCACTTCATAAGTATCG-3´) and FNIb (5´-GCTAAACATTCTGCGTGGATGTAT-3´), R6K1 (5´-CTGGTGTTTAACTTCATGATCCATT-3´)and F6K1 (5´-CGCTCCACACCAGGTGTTAG-3´), RVPg (5´-TTCATGCTCCACTTCCTGTTTTG-3´)and FVPg (5´-GGCAAGAACAAATCCAAAAGAATTC-3´), RCP (5´-CAGTTCTTGACTCCAAGTAGAGTATG-3´)and FCP (5´-GGAAATGACACAATCGATGCAG-3´) (Shen et al. 2018).
Statistical analysis
The EC50 values were determined from concentration-effect curves by linear regression analysis. Statistical analysis was performed using SPSS 20.0, and data were presented as the arithmetic mean ± standard deviation.
Results
Antiviral activities of crude extracts against PVY
As shown in Table 1, both ethyl acetate and n-butanol extracts from strain E303035 showed significant antiviral activity against PVY. The inhibitory activity of ethyl acetate extract was higher than that of n-butanol. The inhibition rates of both extracts were concentration-dependent. The ethyl acetate extract possessed its highest inhibitory activity at 10 mg/mL, with an inhibition rate of 99.70%. The n-butanol extract possessed its highest inhibitory activity at 20 mg/mL, with an inhibition rate of 96.43%. The ethyl acetate extract was then isolated to obtain compounds with anti-viral activity against PVY.
Isolation of compounds from the halophilic bacterium Bacillus pumilus
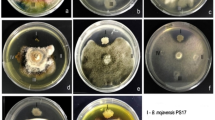
As shown in Fig. 1, 9 compounds were isolated from the ethyl acetate extract of strain E303035 and identified as cyclo(L-Leu-L-Pro) (1) (Yue et al. 2010), cyclo(L-Pro-L-Tyr) (2) (Li et al. 2013), Brevianamide F (3) (Wang et al. 2015), 2-(3-Indolyl) ethanol (4) (Böhlendorf et al. 1996), N-[2-(1H-indol-3-yl) ethyl] acetamide (5) (Anouhe et al. 2015), 3, 3-di(1H-indol-3-yl)propane- 1,2-diol (6) (Zhao et al. 2009), Lincomycin B (7) (Argoudelis et al. 1965), dibutylphthalate (8) (Dai et al. 2006), and p-hydroxyphenethyl alcohol (9) (Lou et al. 2001).
Cyclo(L-Leu-L-Pro) (1). Yield: 86 mg; white amorphous powder; molecular formula C11H18N2O2.
Cyclo(L-Pro-L-Tyr) (2). Yield: 25 mg; colorless oil; molecular formula C14H16N2O3.
Brevianamide F (3). Yield: 11 mg; white powder; molecular formula C16H17N3O2.
2-(3-indolyl) ethanol (4). Yield: 51 mg; white powder; molecular formula C10H11NO.
N-[2-(1H-indol-3-yl) ethyl] acetamide (5). Yield: 78 mg; yellowish-brown oil; molecular formula C12H13N2O.
3, 3-di(1H-indol-3-yl)propane- 1,2-diol (6). Yield: 8 mg; white powder; molecular formula C18H18N2O2.
Lincomycin B (7). Yield: 13 mg; white powder; molecular formula C17H32N2O6S.
Dibutylphthalate (8). Yield: 3900 mg; brownish red oil; molecular formula C16H22O4.
P-hydroxyphenethyl alcohol (9). Yield: 10 mg; colorless needle crystal; molecular formula C8H10O2.
The data of 1H NMR and 13C NMR of compounds 1–9 is in Additional file 1.
Antiviral activities of compounds isolated from strain E303035 against PVY
The antiviral activities, against PVY, of compounds isolated from strain E303035 were determined at 31, 63, 125, 250, and 500 μg/mL. As shown in Table 2, compounds 1, 5, and 9 possessed higher antiviral activity against PVY, with EC50 values of 210.99, 224.26, and 305.37 μg/mL, respectively. Compound 5 showed significant antiviral activity against PVY at 250 and 500 μg/mL, with inhibition values of 62.40% and 75.99%, respectively. Compound 1 also showed moderate antiviral activity at a concentration of 125 μg/mL, with an inhibition value of 53.87%. Compound 4 showed weak antiviral activity at 250 and 500 μg/mL, with inhibition values of 30.21% and 24.29%, respectively. In summary, we found that compounds 1, 5, and 9 had good inhibitory activity against PVY, and compounds 1, 4, 5, and 9 were then used for further research of inhibitory activity against genes encoding functional proteins of PVY.
Effects of active compounds on the expression of genes encoding functional proteins
The expressions of genes HC-pro, P1, P3, NIa, NIb, 6K1, VPg, and CP under treatment with compounds 1, 4, 5, and 9 at 31, 63, 125, 250, and 500 μg/mL were determined by PCR (Figs. 2 and 3).
Effect of active compounds on expression of genes encoding PVY functional proteins: HC-pro, P3, NIb and NIa. a gene HC-Pro; b gene P3; c gene NIb; d gene NIa. M: Marker lane; Series of Lanes 1–5: compound 5; 6–10: compound 1; 11–15: compound 9; 16–20: compound 4. Each series of 5 lanes between markers represents the concentrations used: 500, 250, 125, 63, and 31 μg/mL, from left to right
Effect of active compounds on expression of genes encoded PVY functional proteins of VPg, CP, 6K1, and P1. a gene VPg; b gene CP; c gene 6K1; d gene P1. M: Marker lane; Series of Lanes 1–5: compound 5; 6–10: compound 1; 11–15: compound 9; 16–20: compound 4. Each series of 5 lanes between markers represents the concentrations used: 500, 250, 125, 63, and 31 μg/mL, from left to right
As shown in Fig. 2, organizing data by gene, HC-pro and NIb showed no expression under treatment with compounds 1, 4, and 9 at all concentrations, and with compound 5 at 125 and 500 μg/mL (Fig. 2a, c). Gene P3 showed no expression under treatment with compounds 1, 4, 5, and 9 at all concentrations (Fig. 2b). Gene NIa showed no expression under treatment with compounds 1 and 4 at all tested concentrations, compound 5 at 125 and 500 μg/mL, and compound 9 at 125, 250, and 500 μg/mL (Fig. 2d). Organizing data by compound, at all tested concentrations: 1 and 4 significantly inhibited the expressions of genes HC-pro, P3, NIb, and NIa, compound 5 inhibited gene P3, and compound 9 inhibited NIb.
As shown in Fig. 3, with data organized by gene, genes VPg and 6K1 showed no expression under treatment with compounds 1, 4, 5, and 9 at all tested concentrations (Fig. 3a, c). Gene CP showed no expression under treatment with compound 1 at 63, 125, and 500 μg/mL, compound 4 at 250 and 500 μg/mL, compound 5 at 125 and 500 μg/mL, and compound 9 at 125, 250, and 500 μg/mL (Fig. 3b). Gene P1 showed no expression under treatment with compound 1 at 63, 125, and 500 μg/mL, compound 4 at 63, 250, and 500 μg/mL, compound 5 at 125, 250, and 500 μg/mL, and compound 9 at 63, 125, 250, and 500 μg/mL (Fig. 3d). Organizing the data by compound, compounds 1, 4, 5, and 9 significantly inhibited the expressions of genes VPg and 6K1 at all tested concentrations.
Effect of active compounds on the expression of early response genes induced by potato virus Y
Effects of active compounds on the expression of chlorophyll a-b binding protein (Lhc)
As shown in Fig. 4, the expression of gene Lhc under the treatment of compounds 1, 5, and 9 at 31, 63, 125, 250, and 500 μg/mL after 2, 4, and 6 days were determined by real-time PCR.
For compound 1, increasing the concentration caused significant up-regulation of the gene Lhc. Over time, the gene was significantly down-regulated at day 4, then up-regulated at day 6, at all concentrations. The expression of gene Lhc was at its lowest at day 4 (Fig. 4a).
For compound 5, increasing the concentration caused significant down-regulation of the gene Lhc. The gene was significantly up-regulated at day 4, and then down-regulated at day 6, at all concentrations. The expression of gene Lhc was at its highest at day 4 (Fig. 4b).
For compound 9, increasing the concentration caused significant down-regulation of the gene Lhc. The gene was significantly down-regulated at day 4, and then up-regulated at day 6, at all concentrations. The expression of gene Lhc was at its lowest at day 4 (Fig. 4c).
Effect of active compounds on the expression of photo system II (PSii)
As shown in Fig. 5, the expression of gene PSii under treatments with compounds 1, 5, and 9 at 31, 63, 125, 250, and 500 μg/mL after 2, 4, and 6 days was determined by real-time PCR.
For compounds 1, 5, and 9, increasing the concentration caused significant up-regulation of the gene PSii. The gene was significantly up-regulated at day 4, then down-regulated at day 6, at all concentrations. The expression of gene PSii was at its highest at day 4.
Effect of active compounds on the expression of xyloglucan endotransglucosylase hydrolase (XTH)
As shown in Fig. 6, the expression of gene XTH under treatments with compounds 1, 5, and 9 at 31, 63, 125, 250, and 500 μg/mL after 2, 4, and 6 days was determined by real-time PCR.
For compound 1 and 9, increasing the concentration caused significant down-regulation of the gene XTH. The gene was significantly down-regulated at day 4, and then up-regulated at day 6, at all concentrations. The expression of gene XTH was at its lowest at day 4 (Fig. 6a, c).
For compound 5, increasing the concentration caused significant up-regulation of the gene XTH. The gene was significantly up-regulated at day 4, and then down-regulated at day 6, at all concentrations. The expression of gene XTH was at its highest at day 4 (Fig. 6b).
Effect of active compounds on the expression of gene CP-interacting protein 3 (CPIP3)
As shown in Fig. 7, the expression of gene CPIP3 under treatment with compounds 1, 5, and 9 at 31, 63, 125, 250, and 500 μg/mL after 2, 4, and 6 days was determined by real-time PCR.
For compounds 1 and 5, increasing the concentration caused significant down-regulation of the gene CPIP3. The gene was significantly up-regulated at day 4, and then down-regulated at day 6, at all concentrations. The expression of gene CPIP3 was at its highest at day 4 (Fig. 7a, b).
For compound 9, increasing the concentration caused significant up-regulation of the gene CPIP3. The gene was significantly up-regulated at day 4, and then down-regulated at day 6, at all concentrations. The expression of gene CPIP3 was at its highest at day 4 (Fig. 7c).
Effect of active compounds on the expression of the gene cullin-1 isoform (cullin-1)
As shown in Fig. 8, the expression of gene cullin-1 under treatment with compounds 1, 5, and 9 at 31, 63, 125, 250, and 500 μg/mL after 2, 4, and 6 days was determined by real-time PCR.
For compound 1 and 5, increasing the concentration caused significant down-regulation of the gene cullin-1. The gene was significantly up-regulated at day 4, and then down-regulated at day 6, at all concentrations. The expression of gene cullin-1 was at its highest at day 4 (Fig. 8a, b).
For compound 9, increasing the concentration caused significant up-regulation of the gene cullin-1. The gene was significantly up-regulated at day 4, and then down-regulated at day 6, at all concentrations. The expression of gene cullin-1 was the highest at day 4 (Fig. 8c).
Discussion
The genome of PVY is composed of a single-stranded RNA, length 9.7 kb, which covalently links with a viral-encoded protein (VPg) at its 5ʹ-end and contains a 3ʹ-polyadenylated tail. The genome of PVY encodes two polyproteins, a larger polyprotein of about 3000 amino acids and a shorter one translated from a 2+ frameshift in the P3 coding region. These polyproteins are cleaved by viral proteases, subsequently generating eleven mature proteins (Kim et al. 2015).
Compounds 1 and 4 significantly inhibited the expression of gene HC-pro at all concentrations tested. HC-Pro encodes viral helper component-proteinase (HC-Pro), which is a pathogen elicitor. It is also involved in multiple roles in aphid transmission, RNA binding, suppression of gene silencing and protease activity (Chowdhury et al. 2020; Kumar et al. 2020).
Compounds 1, 4, 5 and 9 significantly inhibited the expression of gene VPg at all concentrations tested. VPg encodes a viral protein called VPg which sabotages host antiviral RNA silencing to promote viral infection (Cheng et al. 2016).
Compounds 1, 4, 5, and 9 significantly inhibited the expression of genes CP and P1, at some concentrations. CP encodes a viral coat protein and P1 encodes plasma membrane cation-binding protein 1 (PCaP1), which is shown to be important for the intra-cellular movement of two members of the genus Potyvirus, in Arabidopsis (rockcress) and in tobacco plants (Beris et al. 2020).
Compounds 1 and 4 significantly inhibited the expression of gene NIa at all concentrations tested. NIa encodes the “nuclear inclusion a” (NIa) protease of PVY, which is involved in processing the polyprotein encoded by its positive sense RNA genome. The NIa protein is also responsible for the release of the VPg protein involved in viral RNA replication (Gargouri-Bouzid et al. 2006).
Compounds 1, 4 and 9 significantly inhibited the expression of gene NIb at all concentrations tested. NIb is an RNA-dependent RNA polymerase in the PVY genome, which has been reported to confer virulence in infections of pepper plants (Janzac et al. 2010).
Compounds 1, 4 and 5 significantly inhibited the expression of gene P3 at all concentrations tested. P3 encodes potyviral membrane protein.
Compounds 1, 4, 5, and 9 significantly inhibited the expression of gene 6K1. The function of 6K1 is currently unknown. In summary, all compounds showed inhibitory activities against various genes encoding functional proteins of PVY. The results provide a starting point for future research into the inhibitory mechanisms and pathways of active compounds against PVY.
The Lhc gene encodes chlorophyll a-b binding protein, which takes part in regulating photosynthesis and chlorophyll synthesis. The down-regulation of this gene means a decrease in viral nucleic acid, which is also seen in PVY-resistant plants (Chen et al. 2017). Compounds 5 and 9 down-regulated the Lhc gene. There was an inverse relationship between concentration and gene expression. At day 4, the expression of gene Lhc was at its lowest. These results suggest that compounds 5 and 9 elicit their inhibitory activity due to their influence on chlorophyll synthesis and photosynthesis during days 2−4.
The PSii gene takes part in photosystem II of photosynthesis. A down-regulation of this gene has the same effect as inhibition of Lhc: a decrease in viral nucleic acid. Compounds 1, 5, and 9 showed no significant inhibitory effect of this gene, but the expression of PSii gene was at its lowest on day 6.
The XTH gene encodes xyloglucan endotransglucosylase hydrolase, which is an essential constituent of the primary cell wall and participates in cell wall elongation and construction (Otulak-Kozieł K et al. 2018a; Otulak-Kozieł K et al. 2018b). The cell wall is the first barrier that protects plants against entry of pathogens and other harmful bodies, and offers protection against mechanical stress to plant cells. The expression of cell-wall-related genes, including XTH, has been shown to be regulated by biotic and abiotic stresses (Li et al. 2005). In this study, XTH was up-regulated under treatment with compound 5 as the concentration increased before 4 days. This suggests that compound 5 could increase elongation and construction of plant cell walls at different concentrations, especially at day 4. However, the XTH gene showed down-regulation under treatment with compounds 1 and 9 at day 4 and then up-regulation at day 6. This suggests that both compounds 1 and 9 could in increase elongation and construction of plant cell walls after 4 days, especially at day 6. It affected the plant cell wall later than that of compound 5. This may indicate that these compounds contribute to induce PVY resistance in Nicotiana.
CPIP3 is a stress response gene, encoding CP-interacting protein 3. The interaction between viral coat protein (CP) and host plant CP-interacting protein were shown to be important for the plant defense response, viral propagation, and long distance movement (Li et al. 2005; Park et al. 2009). A low expression of the CPIP3 gene signifies a low quantity of CP-interacting protein 3 which, to some extent, signifies inhibitory activity against PVY (Li et al. 2005). In this study, compounds 1 and 5 significantly down-regulated the gene CPIP3 at 2 days, with their increased concentration. Therefore, compounds 1 and 5 could inhibit the viral multiplication of PVY by inducing the plant defense response in tobacco plants. Although compound 9 was able to up-regulate CPIP3 at days 2 and 4, by day 6 there was significant down-regulation of the gene, at all concentrations tested. The result demonstrated the inhibitory activity of compound 9 against the multiplication of PVY, by inducing the tobacco plant defense response at 6 days.
Gene cullin-1 also encodes a stress response protein. The up-regulation of it was reported to be important in the resistance response to pathogen infection and autoimmunity (Cheng et al. 2011; Gou et al. 2009; Liu et al. 2002). In our study, the gene cullin-1 was significantly up-regulated by compounds 1, 5, and 9 at days 2 and 4. This meant that all three compounds had significant inhibitory activity against PVY by inducing the resistance response to PVY infection and autoimmunity in tobacco plants at 2–4 days. In detail, compounds 1 and 5 up-regulated cullin-1 at low concentrations of 31, 63, and 125 μg/mL, while compound 9 up-regulated cullin-1 at a high concentration of 500 μg/mL.
Taken together, active compounds 1, 5, and 9 had significant inhibitory activity against PVY and various effects on the expression of viral genes related to pathogenesis. They might be considered as potential antiviral agents for future development.
Availability of data and materials
We admit availability of data and material.
References
Anouhe JBS, Adima AA, Niamké FB, Stien D, Amian BK, Blandinières PA, Virieux D, Pirat JL, Kati-Coulibaly S, Amusant N (2015) Dicorynamine and harmalan-N-oxide, two new b-carboline alkaloids from Dicorynia guianensis Amsh heartwood. Phytochem Let 12:158–163
Argoudelis AD, Fox JA, Eble TE (1965) A new lincomycin-related antibiotic. Biochem 4:698–703
Beris D, Kotsaridis K, Vakirlis N, Termentzi A, Theologidis I, Moury B, Vassilakos N (2020) The plasma membrane Cation binding protein 1 affects accumulation of Potato virus Y in pepper both at the systemic level and in protoplasts. Virus Res. https://doi.org/10.1016/j.virusres.2020.197899
Böhlendorf B, Bedorf N, Jansen R, Trowitzsch-Kienast W, Höfle G, Forche E, Gerth K, Irschik H, Kunze B (1996) Reichenbach H (1996) Antibiotics from gliding bacteria, LXXIII indole and quinoline derivatives as metabolites of tryptophan in Myxobacteria. Euro J Org Chem 1:49–53
Caetano T, Krawczyk JM, Mosker E, Sussmuth RD, Mendo S (2011) Heterologous expression, biosynthesis, and mutagenesis of type II lantibiotics from Bacillus licheniformis in Escherichia coli. Cell Chem Biol 18(1):90–100
Chen S, Li FX, Jiang CH, Cui LJ, Shen LL, Liu GS, Yang AG (2017) Dynamic expression analysis of early response genes induced by potato virus Y in PVY-resistant Nicotiana tabacum. Plant Cell Rep 36:297–311
Cheng XF, Wang AM (2016) The potyvirus silencing suppressor protein VPg mediates degradation of SGS3 via ubiquitination and autophagy pathways. J Virol 91(1):e01478–16
Cheng YT, Li Y, Huang S, Huang Y, Dong X, Zhang Y, Li X (2011) Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc Natl Acad Sci USA 108:14694–14699
Chowdhury RN, Lasky D, Karki H, Zhang Z, Goyer A, Halterman D, Rakotondrafara AM (2020) HCPro suppression of callose deposition contributes to strain-specific resistance against Potato Virus Y. Phytopathol 110(1):164–173
Dai HF, Mei WL, Proksch P, Lin WH (2006) Studies on the tumor cytotoxic constituents from the marine sponge Hyrtios erectus. Chin J Mar Drugs 25:1–5 (Chinese)
Davie K, Holmes R, Pickup J, Lacomme C (2017) Dynamics of PVY strains in field grown potato: impact of strain competition and ability to overcome host resistance mechanisms. Virus Res 241:95–104
Funke CN, Nikolaeva OV, Green KJ, Tran LT, Chikh-Ali M, Quintero-Ferrer A, Cating RA, Frost KE, Hamm PB, Olsen N, Pavek MJ, Gray SM, Crosslin JM, Karasev AV (2017) Strain-specific resistance to Potato virus Y (PVY) in potato and its effect on the relative abundance of PVY strains in commercial potato fields. Plant Dis 101(1):20–28
Gargouri-Bouzid R, Jaoua L, Rouis S, Saïdi MN, Bouaziz D, Ellouz R (2006) PVY-resistant transgenic potato plants expressing an anti-NIa protein scFv antibody. Mol Biotechnol 33:133–140
Gou M, Su N, Zheng J, Huai J, Wu G, Zhao J, He J, Tang D, Yang S, Wang G (2009) An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J 60(5):757–770
Hu HQ, Li XS, He H (2010) Characterization of an antimicrobial material from a newly isolated Bacillus amyloliquefaciens from mangrove for biocontrol of Capsicum bacterial wilt. Bio Control 54(3):359–365
Janzac B, Montarry J, Palloix A, Navaud O, Moury B (2010) A point mutation in the polymerase of Potato virus Y confers virulence toward the Pvr4 resistance of pepper and a high competitiveness cost in susceptible cultivar. Mol Plant Microbe Interact 23(6):823–830
Kim SB, Lee HY, Seo S, Lee JH, Choi D (2015) RNA-dependent RNA polymerase (NIb) of the potyviruses is an avirulence factor for the broad-spectrum resistance gene Pvr4 in Capsicum annuum cv. CM334. PLoS ONE. https://doi.org/10.1371/journal.pone.0119639
Kumar S, Karmakar R, Gupta I, Patel AK (2020) Interaction of potyvirus helper component-proteinase (HcPro) with RuBisCO and nucleosome in viral infections of plants. Plant Physiol Biochem 151:313–322
Li Y, Wu MY, Song HH, Hu X, Qiu BS (2005) Identification of a tobacco protein interacting with tomato mosaic virus coat protein and facilitating long-distance movement of virus. Arch Virol 150(10):1993–2008
Li T, Wang GC, Huang XJ, Ye WC (2013) Whitmanoside A, a new α-pyrone glycoside from the leech Whitmania pigra. Nat Prod 44(44):1537–1843
Liu Y, Schiff M, Serino G, Deng XW, Dinesh-Kumar SP (2002) Role of SCF ubiquitinligase and the COP9 signalosome in the Ngene-mediated resistance response to Tobacco mosaic virus. Plant Cell 14(7):1483–1496
Lou HX, Yuan HQ, Yamazaki Y, Sasaki T, Oka S (2001) Alkaloids and flavonoids from peanut skins. Planta Med 67(4):345–349
Otulak-Kozieł K, Kozieł E, Bujarski JJ (2018a) Spatiotemporal changes in Xylan-1/Xyloglucan and Xyloglucan Xyloglucosyl transferase (XTH-Xet5) as a step-in of ultrastructural cell wall remodelling in potato-potato virus Y (PVYNTN) hypersensitive and susceptible reaction. Int J Mol Sci 19(8):1–23
Otulak-Kozieł K, Kozieł E, Lockhart BEL (2018b) Plant cell wall dynamics in compatible and incompatible potato response to infection caused by Potato Virus Y (PVYNTN). Int J Mol Sci 19(3):1–23
Park MR, Park SH, Cho SY, Kim KH (2009) Nicotiana benthamiana protein, NbPCIP1, interacting with Potato virus X coat protein plays a role as susceptible factor for viral infection. Virol 386:257–269
Shen S, Li W, Wang J (2013) A novel and other bioactive secondary metabolites from a marine fungus Penicillium oxalicum 0312F1. Nat Prod Res 27(24):2286–2291
Shen S, Li W, Ouyang M, Wang J (2018) Structure-activity relationship of triterpenes and derived glycosides against cancer cells and mechanism of apoptosis induction. Nat Prod Res 32(6):654–661
Wang B, Park EM, King JB, Mattes AO, Nimmo SL, Clendinen C, Edison AS, Anklin C, Cichewicz RH (2015) Transferring fungi to a deuterium- enriched medium results in assorted, conditional changes in secondary metabolite production. J Nat Prod 78(6):1415–1421
Xiao JJ, Liao M, Chu MJ, Ren ZL, Zhang X, Lv XH, Cao HQ (2015) Design, synthesis and anti-Tobacco Mosaic Virus (TMV) activity of 5-Chloro-N-(4-cyano-1-aryl-1H-pyrazol-5-yl)-1-aryl-3-methyl- 1H-pyrazole-4-carboxamide derivatives. Molecules 20:807–821
Yue ZG, Zi JC, Zhu CG, Lin S, Yang YC, Shi JG (2010) Constituents of Gymnadenia conopsea. China J Chin Mat Med 35(21):2852–2861 (Chinese)
Zhang J, Zhao L, Zhu C, Wu ZX, Zhang GP, Gan XH, Liu DY, Pan JK, Hu DY, Song BA (2017) Facile synthesis of novel vanillin derivatives incorporating a bis(2-hydroxyethyl)dithhioacetal moiety as antiviral agents. J Agr Food Chem 65(23):4582–4588
Zhao TT, Li XM, Li JM, Li K, Cui CM, Li CS, Wang BG (2009) Chemical constituents of EN-22, an endophytic fungus derived from the marine red alga Polysiphonia urceolata. Mar Sci 33(7):81–86 (Chinese)
Acknowledgements
We thank Prof. Bao-An Song and his staff in Guizhou University for technical assistance.
Funding
This research was financed by the Natural Science Fund of Qinghai Province (2019-ZJ-914) and National Natural Science Fund (31860512).
Author information
Authors and Affiliations
Contributions
WL and SS conceived and designed research. WL and SS conducted experiments. SS carried out all experiments. WL and SS analyzed data. SS wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
This article does not contain any studies with human participants or animals. We declare consent to participate.
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
The data of 1H NMR and 13C NMR of compounds 1–9
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shen, S., Li, W. The inhibitory effects of metabolites from Bacillus pumilus on potato virus Y and the induction of early response genes in Nicotiana tabacum. AMB Expr 10, 152 (2020). https://doi.org/10.1186/s13568-020-01089-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-020-01089-1











