Abstract
Background
Gestational age at delivery is associated with health and social outcomes. Recently, cord blood DNA methylation data has been used to predict gestational age. The discrepancy between gestational age predicted from DNA methylation and determined by ultrasound or last menstrual period is known as gestational age acceleration. This study investigated associations of sex, socioeconomic status, parental behaviours and characteristics and birth outcomes with gestational age acceleration.
Results
Using data from the Avon Longitudinal Study of Parents and Children (n = 863), we found that pre-pregnancy maternal overweight and obesity were associated with greater gestational age acceleration (mean difference = 1.6 days, 95% CI 0.7 to 2.6, and 2.9 days, 95% CI 1.3 to 4.4, respectively, compared with a body mass index < 25 kg/m2, p < .001). There was evidence of an association between male sex and greater gestational age acceleration. Greater gestational age acceleration was associated with higher birthweight, birth length and head circumference of the child (mean differences per week higher gestational age acceleration: birthweight 0.1 kg, 95% CI 0.1 to 0.2, p < .001; birth length 0.4 cm, 95% CI 0.2 to 0.7, p < .001; head circumference 0.2 cm, 95% CI 0.1 to − 0.4, p < .001). There was evidence of an association between gestational age acceleration and mode of delivery (assisted versus unassisted delivery, odds ratio = 0.9 per week higher gestational age acceleration, 95% CI 0.7, 1.3 (p = .05); caesarean section versus unassisted delivery, odds ratio = 0.6, 95% CI 0.4 to 0.9 per week higher gestational age acceleration (p = .05)). There was no evidence of association for other parental and perinatal characteristics.
Conclusions
The associations of higher maternal body mass index and larger birth size with greater gestational age acceleration may imply that maternal overweight and obesity is associated with more rapid development of the fetus in utero. The implications of gestational age acceleration for postnatal health warrant further investigation.
Similar content being viewed by others
Background
Preterm birth (≤ 37 weeks gestation) is associated with numerous health consequences such as increased mortality [1], hypertension [2, 3], insulin resistance [4] and respiratory problems in later life [5, 6]. Indeed, each additional gestational day at birth is associated with improved medical and neuropsychological outcomes in childhood [7]. Gestational age (GA) at delivery is typically determined via early obstetric ultrasound or last menstrual period (LMP), with ultrasound methods considered the more reliable [8], ‘gold standard’ procedure.
Recently, DNA methylation (DNAm) has been used to predict GA at delivery [9, 10]. This method builds on work that used DNAm to predict chronological age [11] and subsequent work, showing that the differences between predicted and chronological age are associated with disease outcomes. Those with DNAm-predicted ages that exceeded their chronological ages (age acceleration, AA) have a higher risk of cancer incidence [12,13,14], Alzheimer’s disease [15] and mortality [15,16,17,18,19,20]. Of note, the term AA is commonly used in the literature to describe both positive and negative differences (i.e. predicted ages above or below chronological ages), which could be misleading, but we use the term to be consistent with previous literature.
In a similar way, the gestational epigenetic clocks developed by Bohlin et al. [10] and Knight et al. [9] can be compared with actual GA to determine gestational age acceleration (GAA). There is little existing research on the association of potential predictors of GAA or the potential outcomes of GAA.
The aim of this study was to apply a previously published model for predicting GA from DNAm [10] and use this model to estimate GAA in order to (i) explore potential predictors of GAA by assessing the association of a broad range of socioeconomic variables and parental characteristics with GAA and (ii) explore potential outcomes of GAA by assessing associations of GAA with delivery and postnatal factors, using data from the Accessible Resource for Integrated Epigenomic Studies (ARIES) project, a subsample of child-mother pairs from the Avon Longitudinal Study of Parents and Children (ALSPAC).
Results
Study sample characteristics
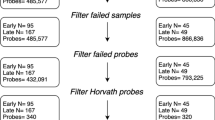
The characteristics of the 863 participants from the ARIES cohort included in our analysis are displayed in Tables 1 and 2, and Additional file 1: Table S1 describes the differences between these participants and the full ALSPAC cohort from which ARIES is a subsample. Additional file 1: Table S2 shows the full range of GAs of the 863 participants included in the analysis.
Modelling of GA estimates
GA was estimated from ARIES cord blood DNAm using the model of Bohlin et al. [10]. Correlation between estimated and reported GA was high (correlation coefficient r = 0.65) though not as high as that reported in the original publication (r = 0.81). Some reduction, however, was expected because the Bohlin et al. [10] model was trained and tested in distinct subsets of the same Norwegian cohort. We elected not to use the model of Knight et al. [9] because we were less confident that it would produce meaningful GAA estimates given its low correlation with GA in ARIES (r = 0.37) [21].
Associations of gender, socioeconomic and parental factors with GAA
Females had lower GAA than males by 0.8 days after adjusting for sex and cell type proportion (mean difference [MD] = − 0.8 days; 95% CI − 1.4, − 0.1, p = .024; Table 3).
Maternal pre-pregnancy overweight and obese status was associated with higher GAA compared with maternal pre-pregnancy body mass index (BMI) of < 25 kg/m2 (MD = 1.6 days, 95% CI 0.7, 2.6 for overweight; MD = 2.9 days, 95% CI 1.3, 4.4 for obese, p < .001, see Table 4) after adjusting for sex, cell type proportion and parental socioeconomic factors. As GAA calculated using Bohlin et al. methods [10] is correlated with birthweight [21], we further adjusted these models including birthweight as a covariate to assess whether birthweight may be driving this effect. The results were not substantially different when additionally adjusting for birthweight (Additional file 1: Table S3).
No clear associations were found for parental education, relationship status, smoking, alcohol consumption, depression or age, nor with housing tenure, financial difficulties, parity or pregnancy complications with GAA (Tables 3 and 4).
Associations of GAA with delivery and birth outcomes
There were strong positive associations of GAA with birthweight (MD = 0.1 kg of birthweight per week higher GAA, 95% CI 0.1, 0.2, p < .001), birth length (MD = 0.4 cm of birth length per week higher GAA, 95% CI 0.2, 0.7, p < .001) and head circumference (MD = 0.2 cm of head circumference per week higher GAA, 95% CI 0.1, 0.4, p < .001) (see Table 5).
No association was observed between GAA and Appearance, Pulse, Grimace, Activity, and Respiration (APGAR) score at 5 min. GAA was inversely associated with assisted delivery (excluding caesarean section) (odds ratio [OR] = 0.9 per week higher GAA, 95% CI 0.7, 1.3, see Table 5) and caesarean section (OR = 0.6 per week higher GAA, 95% CI 0.4, 0.9, p = .05).
There were no substantial differences between the imputed and observed data sets (Additional file 1: Table S4). Adjusting for cell type proportion did not substantially alter our results (see Additional file 1: Tables S5 to S7 for unadjusted results) and neither did adjustment for socioeconomic status (SES) and other potential confounders.
Discussion
Our analyses indicated that male sex, higher maternal pre-pregnancy BMI and vaginal delivery are associated with higher GAA. Our results also indicated that higher GAA was associated with birth size (birthweight, birth length and head circumference). There was no clear evidence of any associations of GAA with parental education, relationship status, smoking, alcohol consumption, depression or age, nor with housing tenure, financial difficulties, parity, pregnancy complications or APGAR scores.
Unlike in AA research, where AA has been associated with negative outcomes such as all-cause mortality and Alzheimer’s disease [15,16,17,18,19,20], it is currently unclear whether accelerated GA at birth is beneficial or detrimental to a fetus or newborn. From previous research using ARIES data, we know sex, birthweight, caesarean section and maternal BMI have also been associated with AA [22]. A recent study has also shown associations of GAA with birth size and sex [23]. However, in their main analysis, associations with birth size were in the opposite direction to our analysis when the raw GAA-GA difference was used as the outcome, i.e. higher GAA was associated with smaller size at birth. When calculating GAA using the residuals from a regression of DNAm-predicted GA on reported GA, as GAA was calculated in this study, there was no clear association between GAA and size at birth. In further contrast, we did not replicate their associations with maternal age, APGAR scores (at 1 min) and pregnancy complications (pre-eclampsia). The discrepancies in the results could be related to the focus on raw differences rather than residuals in the main analysis of Girchenko et al. [23]. We did not estimate GAA as the difference between DNAm-predicted GA and reported GA because the confounding effect of GA is not accounted for in this approach, whereas the residual-based approach ensures GAA is uncorrelated with GA.
Another explanation for the discrepancies between our findings and the findings of Girchenko et al. [23] is their use of the Knight et al. [9] GA prediction model rather than the Bohlin et al. [10] model applied in this study. We have previously noted several key methodological differences in the derivation of the Knight et al. [9] and the Bohlin et al. [10] models that influence the accuracy of the GA prediction in this cohort [21], such as the inclusion of preterm infants in the test set of the Knight et al. model [9] which is inappropriate for a data set with few pre-term births (as in this study). Additionally, the number of CpGs (148) included in the Knight et al. [9] training model was close to the sample size of the training set (207) and the model was then tested with a much larger sample size which may have resulted in an overfitting of the model. In contrast, the number of CpGs (96) in the Bohlin et al. [10] training model was much lower than the sample size of the training set (1068). Thus, the Bohlin et al. [10] model provided us with the best estimate of GAA for this cohort [21], which was reflected in the stronger correlation found between reported GA in the ARIES cohort and the model predictions of GA compared to the Knight et al. [9] model. The Bohlin et al. [10] model for prediction of GA from DNAm performed well in our data, adding support to the notion that DNAm could potentially be used as a marker of GA in data sets where GA has not been measured.
In this study, we were able to apply the epigenetic clock created by Bohlin et al. [10] in ARIES data. Despite the original model being trained only on a Norwegian cohort, the methods of Bohlin et al. [10] transferred to a UK cohort with considerable accuracy (r = 0.65). Additionally, the use of ARIES and ALSPAC data (a large and rich source of longitudinal data for children and their families) allowed us to assess the associations of GAA with a wide range of socioeconomic, parental and perinatal factors. The longitudinal nature of ARIES also enabled comparisons between GAA and AA at multiple ages.
Although the ARIES sub-sample is a more affluent sample of the full ALSPAC cohort, our results were robust to adjustment for SES. This is in line with some evidence that such differences are unlikely to severely bias association studies [24,25,26,27]. To account for missing data, we used multiple imputation, which maximised our statistical power. The results were consistent when using complete-case observed data.
Following Gervin et al.’s [28] methods, the regression analyses were adjusted for cell type proportions, even though GA is associated with variation in cell type proportions and this adjustment could therefore potentially bias the results. However, comparing the data before and after adjustment, there were no substantial differences in the results. Additionally, only weak associations were found between GA and cell type composition in this study (Additional file 1: Figure S1). Another potential issue with adjusting for cell type composition arises from deriving cord blood references from full-term births only; cord blood cell counts may be inaccurate for cord blood methylation profiles of preterm infants. However, a small proportion of the participants in our study were pre-term (3%) so it is unlikely that this will have substantially impacted the results.
Interestingly, there is little overlap between probes used in the Bohlin et al. [10] model of GA predication and the Horvath [11] model of age prediction, with only one CpG site (cg08965235 in the latent-transforming growth factor beta-binding protein 3 gene) overlapping between the models. In contrast to the Horvath [11] model, which compares accurate measures of chronological age and epigenetic predictions, the Bohlin et al. [10] model compares potentially inaccurate LMP/ultrasound estimates to epigenetic predictions. Consequently, inaccurate GA estimates may have impacted upon the GAA estimates in this research, especially as the majority of the GA estimates in ALSPAC are derived from LMP, since ultrasound estimation of GA was not common at the time of recruitment. Additionally, the GA predictions using the Bohlin et al. [10] model were more accurate using ultrasound methods rather than LMP. This may mean that our estimates may not be as accurate as if ultrasound GAs had been used and this could explain the discrepancy in accuracy between the Bohlin et al. [10] test data and the ARIES data, as detailed by Simpkin and colleagues [21].
Conclusions
Our results suggest that higher maternal BMI is strongly associated with higher GAA and that higher GAA is strongly associated with larger size at birth (birthweight, birth length and head circumference). In addition, we found weaker associations of sex and delivery method with GAA. Our results may indicate that having a BMI over 25 kg/m2 is associated with more rapid development of the fetus in utero. The implications of GAA for postnatal growth, development and health warrant further investigation.
Methods
Study population
This study used DNAm data generated under the auspices of the ALSPAC [29, 30]. ALSPAC recruited 14,541 pregnant women with expected delivery dates between April 1991 and December 1992. Of these initial pregnancies, there were 14,062 live births and 13,988 children who were alive at 1 year of age. The study website contains details of all the data that are available through a fully searchable data dictionary (http://www.bris.ac.uk/alspac/researchers/data-access/data-dictionary).
As part of the ARIES [31] project (http://www.ariesepigenomics.org.uk), a sub-sample of 1018 ALSPAC child-mother pairs had DNAm measured using the Infinium HumanMethylation450 BeadChip (Illumina, Inc.) [32]. The ARIES sub-sample was selected based on availability of DNA samples at three time points (birth, mean 7.5 years and mean 15.5 years). DNAm was measured three times in ALSPAC offspring, from cord blood at birth and from peripheral blood at approximately ages 7 and 17.
Laboratory methods, quality control and pre-processing
All DNAm wet-lab and pre-processing analyses were performed at the University of Bristol as part of the ARIES project. Following extraction, DNA was bisulphite converted using the Zymo EZ DNA MethylationTM kit (Zymo, Irvine, CA). Infinium HumanMethylation450 BeadChips were used to measure genome-wide DNAm levels at over 485,000 CpG sites. The arrays were scanned using an Illumina iScan, with initial quality review using GenomeStudio. The level of DNAm is expressed as a ‘beta’ value (β value), ranging from 0 (no cytosine methylation) to 1 (complete cytosine methylation). β values are reported as percentages. Several quality control steps were included in the laboratory pipeline which are described in detail elsewhere [33].
Epigenetic GA prediction
Using a recently published model [10], we derived epigenetic gestational age (EGA) from cord blood DNAm. The Bohlin et al. [10] model was chosen over the Knight et al. [9] model due to its much stronger correlation with GA in ARIES (r = 0.65 compared to r = 0.37). This epigenetic clock for GA at delivery uses 96 CpG sites to predict GA from cord blood methylation. We obtained GAA as the residuals from a regression of EGA on observed GA. GA was gathered from clinical records and determined by LMP for the majority; however, on some occasions, this measure was updated following a dating ultrasound. It is not known for which individual GA was based on LMP or ultrasound but as updating GA based on ultrasound was not common practice at the time of the measurement, the numbers are likely to be low. To be consistent with previous literature, we have used the terms ‘AA’ and ‘GAA’ to describe both positive and negative differences (i.e. predicted ages above or below chronological/gestational ages). A positive GAA corresponds to an EGA that was higher than actual GA and vice versa.
Socioeconomic, parental and perinatal characteristics
Socioeconomic factors included housing tenure, social class, parental education and financial difficulties. Parental factors included parental smoking, alcohol use, mental health, relationship status, BMI and age. Finally, perinatal variables considered were child’s sex, birthweight, birth length, head circumference and APGAR score at 5 min as well as the occurrence of any pregnancy complications and the delivery type. All variables were measured through questionnaires at different times during pregnancy (socioeconomic and parental variables), by trained ALSPAC staff shortly after birth (anthropometry at birth) or from obstetric records (pregnancy complications, child’s sex and APGAR score). Full details of measurement of these factors are in Additional file 1.
Statistical analysis
Sex, SES, parental behaviours and pregnancy complications were analysed as potential determinants of GAA. Associations between these factors and GAA were assessed in linear regression models with GAA as the outcome. Models with parental behaviours as the exposure were adjusted for SES variables as confounders. Birth size, delivery method and APGAR score were considered as potential outcomes of GAA. Associations of GAA with these factors were assessed using linear or multinomial logistic regression as appropriate, with GAA as an exposure and parental behaviour and SES variables included as potential confounders. We performed the analysis in this way due to the temporal ordering of the variables, although we do not necessarily hypothesise a direct causal effect of GAA on these outcome variables. The associations were analysed in two models: (1) adjusted for sex and cell type proportion and (2) with additional adjustment for potential confounders, as appropriate for the specific model. The Gervin et al. [28] methods were used for cell-type proportion estimations. Due to missingness in the observed data set, analyses were completed using 100 multiply imputed data sets (see Additional file 1: Table S8 for further information). There were no substantial differences between the analysis of the observed data and the multiply imputed data sets.
Abbreviations
- AA:
-
Age acceleration
- ALSPAC:
-
Avon Longitudinal Study of Parents and Children
- APGAR:
-
Appearance, Pulse, Grimace, Activity, and Respiration
- ARIES:
-
Accessible Resource for Integrated Epigenomic Studies
- BMI:
-
Body mass index
- DNAm:
-
DNA methylation
- EGA:
-
Epigenetic gestational age
- EPDS:
-
Edinburgh Postnatal Depression Scale
- GA:
-
Gestational age
- GAA:
-
Gestational age acceleration
- HA:
-
Owned by the housing association
- LMP:
-
Last menstrual period
- MD:
-
Mean differences
- OR:
-
Odds ratio
- SD:
-
Standard deviation
- SES:
-
Socio-economic status
References
Field D, Draper ES, Fenton A, Papiernik E, Zeitlin J, Blondel B, Cuttini M, Maier RF, Weber T, Carrapato M, et al. Rates of very preterm birth in Europe and neonatal mortality rates. Arch Dis Child-Fetal. 2009;94:F253–6.
de Jong F, Monuteaux MC, van Elburg RM, Gillman MW, Belfort MB. Systematic review and meta-analysis of preterm birth and later systolic blood pressure. Hypertension. 2012;59:226–34.
Evensen KA, Steinshamn S, Tjonna AE, Stolen T, Hoydal MA, Wisloff U, Brubakk AM, Vik T. Effects of preterm birth and fetal growth retardation on cardiovascular risk factors in young adulthood. Early Hum Dev. 2009;85:239–45.
Tinnion R, Gillone J, Cheetham T, Embleton N. Preterm birth and subsequent insulin sensitivity: a systematic review. Arch Dis Child. 2014;99:362–8.
Pike KC, Lucas JS. Respiratory consequences of late preterm birth. Paediatr Respir Rev. 2015;16:182–8.
Kotecha SJ, Edwards MO, Watkins WJ, Henderson AJ, Paranjothy S, Dunstan FD, Kotecha S. Effect of preterm birth on later FEV1: a systematic review and meta-analysis. Thorax. 2013;68:760–6.
Baron IS, Litman FR, Ahronovich MD, Baker R. Late preterm birth: a review of medical and neuropsychological childhood outcomes. Neuropsychol Rev. 2012;22:438–50.
Hoffman CS, Messer LC, Mendola P, Savitz DA, Herring AH, Hartmann KE. Comparison of gestational age at birth based on last menstrual period and ultrasound during the first trimester. Paediatr Perinat Ep. 2008;22:587–96.
Knight AK, Craig JM, Theda C, Baekvad-Hansen M, Bybjerg-Grauholm J, Hansen CS, Hollegaard MV, Hougaard DM, Mortensen PB, Weinsheimer SM, et al. An epigenetic clock for gestational age at birth based on blood methylation data. Genome Biol. 2016;17:206.
Bohlin J, Haberg SE, Magnus P, Reese SE, Gjessing HK, Magnus MC, Parr CL, Page CM, London SJ, Nystad W. Prediction of gestational age based on genome-wide differentially methylated regions. Genome Biol. 2016;17:207.
Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115.
Zheng Y, Joyce BT, Colicino E, Liu L, Zhang W, Dai Q, Shrubsole MJ, Kibbe WA, Gao T, Zhang Z, et al. Blood epigenetic age may predict cancer incidence and mortality. Cancer Res. 2016;76:68–73.
Lin Q, Wagner W. Epigenetic aging signatures are coherently modified in cancer. PLoS Genet. 2015;11:e1005334.
Levine ME, Hosgood HD, Chen B, Absher D, Assimes T, Horvath S. DNA methylation age of blood predicts future onset of lung cancer in the women’s health initiative. Aging-Us. 2015;7:690–700.
Levine ME, Lu AT, Bennett DA, Horvath S. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer’s disease related cognitive functioning. Aging-Us. 2015;7:1198–211.
Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, Gibson J, Henders AK, Redmond P, Cox SR, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25.
Christiansen L, Lenart A, Tan QH, Vaupel JW, Aviv A, McGue M, Christensen K. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2016;15:149–54.
Horvath S, Pirazzini C, Bacalini MG, Gentilini D, Di Blasio AM, Delledonne M, Mari D, Arosio B, Monti D, Passarino G, et al. Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging-Us. 2015;7:1159–70.
Marioni RE, Shah S, Mcrae AF, Ritchie SJ, Muniz-Terrera G, Harris SE, Gibson J, Redmond P, Cox SR, Pattie A, et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol. 2015;44:1388–96.
Breitling LP, Saum K, Perna L, Schöttker B, Holleczek B, Brenner H. Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin Epigenetics. 2016;8:21.
Simpkin AJ, Suderman M, Howe LD. Epigenetic clocks for gestational age: statistical and study design considerations. Clin Epigenetics. 2017;9:100.
Simpkin AJ, Hemani G, Suderman M, Gaunt TR, Lyttleton O, McArdle WL, Ring SM, Sharp GC, Tilling K, Horvath S, et al. Prenatal and early life influences on epigenetic age in children: a study of mother-offspring pairs from two cohort studies. Hum Mol Genet. 2016;25:191–201.
Girchenko P, Lahti J, Czamara D, Knight AK, Jones MJ, Suarez A, Hamalainen E, Kajantie E, Laivuori H, Villa PM, et al. Associations between maternal risk factors of adverse pregnancy and birth outcomes and the offspring epigenetic clock of gestational age at birth. Clin Epigenetics. 2017;9:49.
Osler M, Kriegbaum M, Christensen U, Holstein B, Nybo Andersen AM. Rapid report on methodology: does loss to follow-up in a cohort study bias associations between early life factors and lifestyle-related health outcomes? Ann Epidemiol. 2008;18:422–4.
Greene N, Greenland S, Olsen J, Nohr EA. Estimating bias from loss to follow-up in the Danish National Birth Cohort. Epidemiology. 2011;22:815–22.
Howe LD, Tilling K, Galobardes B, Lawlor DA. Loss to follow-up in cohort studies bias in estimates of socioeconomic inequalities. Epidemiology. 2013;24:1–9.
Winding TN, Andersen JH, Labriola M, Nohr EA. Initial non-participation and loss to follow-up in a Danish youth cohort: implications for relative risk estimates. J Epidemiol Community Health. 2014;68:137–44.
Gervin K, Page CM, Aass HCD, Jansen MA, Fjeldstad HE, Andreassen BK, Duijts L, van Meurs JB, van Zelm MC, Jaddoe VW, et al. Cell type specific DNA methylation in cord blood: a 450K-reference data set and cell count-based validation of estimated cell type composition. Epigenetics. 2016;11:690–8.
Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, Molloy L, Ness A, Ring S, Smith GD. Cohort profile: the ‘children of the 90s’—the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2013;42:111–27.
Fraser A, Macdonald-Wallis C, Tilling K, Boyd A, Golding J, Smith GD, Henderson J, Macleod J, Molloy L, Ness A, et al. Cohort profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol. 2013;42:97–110.
Relton CL, Gaunt T, McArdle W, Ho K, Duggirala A, Shihab H, Woodward G, Lyttleton O, Evans DM, Reik W. Data resource profile: accessible resource for integrated epigenomic studies (ARIES). Int J Epidemiol. 2015;44:1181–90.
Dedeurwaerder S, Defrance M, Calonne E, Denis H, Sotiriou C, Fuks F. Evaluation of the Infinium Methylation 450K technology. Epigenomics. 2011;3:771–84.
Simpkin AJ, Suderman M, Gaunt TR, Lyttleton O, McArdle WL, Ring SM, Tilling K, Smith GD, Relton CL. Longitudinal analysis of DNA methylation associated with birth weight and gestational age. Hum Mol Genet. 2015;24:3752–63.
Acknowledgements
We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. This publication is the work of the authors, and JNK and LDH will serve as guarantors for the contents of this paper.
Funding
This work was supported by grant from the UK Economic and Social Research Council [ES/N000382/1, ES/N000498/1]. LDH is supported by a Career Development Award fellowship from the UK Medical Research Council [MR/M020894/1]. LMOK is supported by a UK Medical Research Council Population Health Scientist fellowship [MR/M014509/1]. JNK, AJA, LMOK, KHW, LCH, CLR, MS and LDH work in a unit that receives funding from the University of Bristol and the UK Medical Research Council [MC_UU_12013/2, MC_UU_12013/3, MC_UU_12013/4, MC_UU_12013/6, MC_UU_12013/9]. The UK Medical Research Council and the Wellcome Trust [grant ref.: 102215/2/13/2] and the University of Bristol provide core support for ALSPAC. JNK is a PhD student funded by the MRC Integrative Epidemiology Unit at Bristol University. ARIES was funded by the BBSRC (BBI025751/1 and BB/I025263/1). Supplementary funding to generate DNAm data which is (or will be) included in ARIES has been obtained from the MRC, ESRC, NIH and other sources. ARIES is maintained under the auspices of the MRC Integrative Epidemiology Unit at the University of Bristol (MC_UU_12013/2 and MC_UU_12013/8).
Availability of data and materials
The data sets generated and/or analysed during the current study are available from the corresponding author on reasonable request, subject to the ALSPAC study executive data access procedures, as specified on the ALSPAC website (http://www.bristol.ac.uk/alspac/researchers/access/) for researchers who meet the criteria for access to confidential data.
Author information
Authors and Affiliations
Contributions
The study was conceived and designed by AJS and LDH. Statistical analyses were conducted by JNK and AJS. JNK was responsible for writing the first draft of the manuscript. All authors contributed to the interpretation of results and critical revisions of the manuscript and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees. Informed consent was given by the offsprings’ parents at the outset of the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Tables S1–S8. Supplementary methods. Details of measurement of socioeconomic, family and perinatal variables and supplementary tables. (DOCX 671 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Khouja, J.N., Simpkin, A.J., O’Keeffe, L.M. et al. Epigenetic gestational age acceleration: a prospective cohort study investigating associations with familial, sociodemographic and birth characteristics. Clin Epigenet 10, 86 (2018). https://doi.org/10.1186/s13148-018-0520-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13148-018-0520-1