Abstract
Background
Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi are common enteric pathogens in humans and animals. Data on the transmission of these pathogens are scarce from Guangdong, China, which has a subtropical monsoon climate and is the epicenter for many emerging infectious diseases. This study was conducted to better understand the prevalence and identity of the three pathogens in pre-weaned dairy calves in Guangdong.
Methods
The occurrence and genetic identity of three pathogens were analyzed by polymerase chain reaction. PCR-positive products were sequenced to determine the species and genotypes. A Chi-square test was used to compare the prevalence of pathogens among sampling dates, age groups, or clinical signs.
Results
The detection rates of Cryptosporidium spp., G. duodenalis and E. bieneusi were 24.0% (93/388), 74.2% (288/388) and 15.7% (61/388), respectively. Three Cryptosporidium species were detected, including C. bovis (n = 73), C. parvum (n = 12) and C. ryanae (n = 7); one animal had concurrence of C. bovis and C. parvum. C. parvum was the dominant species during the first two weeks of life, whereas C. bovis and C. ryanae were mostly seen at 3–9 weeks of age. Sequence analysis identified the C. parvum as subtype IIdA19G1. Assemblage E (n = 282), assemblage A (n = 1), and concurrence of A and E (n = 5) were identified among G. duodenalis-positive animals using multilocus genotyping (MLG). Altogether, 15, 10 and 17 subtypes of assemblage E were observed at the bg, gdh and tpi loci, respectively, forming 49 assemblage E MLGs. The highest detection rate of G. duodenalis was found in winter. Sequence analysis identified genotypes J (n = 57), D (n = 3) and one concurrence of J and D among E. bieneusi-positive animals. The detection rate of E. bieneusi was significantly higher in spring (38.0%; 41/108) than in summer (7.2%; 8/111) and winter (7.1%; 12/169).
Conclusions
These results indicate a common occurrence of C. parvum subtype IIdA19G1, G. duodenalis assemblage E, and E. bieneusi genotype J in pre-weaned dairy calves in Guangdong. More studies are needed to understand the unique genetic characteristics and zoonotic potential of the three enteric pathogens in the province.
Similar content being viewed by others
Background
Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi are common zoonotic pathogens, causing diarrhea and other gastrointestinal illness in humans and animals and responsible for significant morbidity and mortality. In humans, they are transmitted by direct contact with infected persons (anthroponotic transmission) or animals (zoonotic transmission) or through consumption of contaminated food or water (food-borne or water-borne transmission) [1,2,3,4].
In case-control studies, contact with cattle has been implicated as a major risk factor in the epidemiology of human cryptosporidiosis [5]. A wide range of Cryptosporidium species have been identified in cattle, but studies worldwide have shown a common occurrence of C. parvum, C. bovis, C. ryanae and C. andersoni [3]. However, the prevalence of C. parvum in dairy cattle appears to be different between China and other countries [4, 6]. In China, pre-weaned dairy calves are mainly infected with C. bovis rather than C. parvum [7,8,9,10,11,12,13], although concurrence of C. parvum is increasingly detected [14,15,16,17,18]. Elsewhere in the world, C. parvum is responsible for over 90% of the infections in pre-weaned dairy calves [3]; only a few studies in Sweden, Egypt and Malaysia [19,20,21] showed C. bovis being relatively more common.
Subtyping of C. parvum targeting the 60-kD glycoprotein (gp60) gene identified mainly IIa and IId subtypes in dairy cattle [3]. Characterizations of C. parvum isolates in most industrialized countries of America, Europe and Asia suggest that pre-weaned dairy calves are almost exclusively infected with IIa subtypes, with IIaA15G2R1 as the most common subtype [3] while IId subtypes are only seen in Sweden, Egypt and Malaysia, where C. bovis is also commonly co-occurring in pre-weaned dairy calves [19,20,21]. In contrast, IId subtypes are common in pre-weaned calves in China, including IIdA14G1, IIdA15G1, IIdA17G1, IIdA19G1 and IIdA20G1 [10, 11, 13, 15, 16, 18]. Among them, IIdA15G1 and IIdA19G1 are the two major subtypes, responsible for over 90% of C. parvum infections [6].
Thus far, at least eight assemblages (A-H) of G. duodenalis have been identified. Among them, assemblages A and B have been found in humans and many other mammals whereas the remaining ones are mostly host-specific [2]. Most studies in Europe, America, Asia, Australia and Africa identified assemblage E as the genotype responsible for the majority of G. duodenalis infections in pre-weaned dairy cattle [12, 13, 17, 22,23,24,25,26,27,28,29,30], although in a few studies assemblages A and B were detected [31, 32]. Within assemblage A, there are two common sub-assemblages, AI and AII. The former is more commonly found in animals, whereas the latter is more commonly found in humans [2]. A few recent studies have also identified assemblage E in humans [33,34,35].
Over 250 E. bieneusi genotypes have been detected worldwide, including > 40 genotypes in cattle. They mostly belong to nine groups. Among the genotypes in cattle, at least 15, including eight genotypes in group 1 and seven genotypes in group 2, have been reported in humans [36,37,38], revealing the zoonotic potential of bovine E. bieneusi genotypes. The most prevalent genotypes in pre-weaned dairy calves worldwide are I, J and BEB4, all within group 2 [17, 27, 37, 39,40,41,42].
Pre-weaned dairy calves are the most common hosts of zoonotic Cryptosporidium, G. duodenalis and E. bieneusi genotypes. In China, these pathogens have been detected in pre-weaned dairy calves in many areas [8,9,10,11,12,13, 16,17,18, 24, 25, 27, 29, 32, 39,40,41, 43,44,45,46]. Guangdong is the epicenter of many emerging infectious diseases, and has intensive dairy farming and a subtropical monsoon climate with a warm and rainy spring, a long, hot and humid summer, and a mild and dry winter; the mean temperature in spring, summer and winter in Zhaoqing is about 22, 32 and 12 °C, respectively. Data on the prevalence of these pathogens, however, are scarce from Guangdong. Thus far, there has been only one recent study on molecular epidemiology of Cryptosporidium spp. in dairy cattle in the province, which identified C. bovis, C. ryanae and C. andersoni in pre-weaned dairy calves. There has also only been one study on G. duodenalis in the province, which reported a positive rate of 8.4% and identified the presence of assemblage E [47]. No studies have been conducted on E. bieneusi infections in pre-weaned dairy calves in this province. As C. parvum is still relatively uncommon in dairy cattle in China [4], and this zoonotic species has thus far not been reported in Guangdong, there is a need for more studies on the occurrence of C. parvum, G. duodenalis and E. bieneusi in pre-weaned dairy calves in this province.
The aim of the present study was to further examine the prevalence and identity of Cryptosporidium spp., G. duodenalis and E. bieneusi in pre-weaned dairy calves in Guangdong.
Methods
Specimen collection
A total of 388 specimens were collected from pre-weaned Holstein calves in Zhaoqing, Guangdong from March 2017 to January 2018, corresponding to spring, summer and winter seasons in the northern hemisphere. The farm that participated in this study is 0.7 km2 in area and had ~5000 animals at the time of the initial sampling, of which about 400 were calves. This farm adopts the intensive dairy farming system commonly seen in modern dairy operations. The cattle in the farm were grouped according to age, and all newborn calves and pre-weaned calves were kept separately from the older dairy herd. All pre-weaned calves were kept in separate stalls, which were placed adjacent to each other.
In this cross-sectional study, all pre-weaned calves on the farm were sampled on one of the three sampling dates. As there was an approximately 3-month interval between samplings, each animal participated in the study was sampled only once. Approximately 25 g fresh fecal material was collected from the rectum of each calf into 50-ml centrifuge tubes, transported to the laboratory in coolers with ice packs, and stored in 2.5% potassium dichromate at 4 °C before DNA extraction [48].
DNA extraction
After being washed three times with distilled water by centrifugation, about 250 μl of the concentrated fecal material was used in DNA extraction with the Fast DNA SPIN Kit for Soil (MP Biomedical, Santa Ana, CA, USA), following the manufacturer-recommended procedure. DNA, eluted in 100 μl reagent-grade water, was stored at -20 °C before PCR analysis.
PCR analysis
For the detection of Cryptosporidium spp., a 587-bp fragment of the small subunit (SSU) rRNA gene was amplified by nested PCR [49]. Cryptosporidium species presented were genotyped by DNA sequence analysis of positive PCR products. All C. parvum-positive specimens were subtyped by PCR and sequence analysis of an ~850-bp fragment of the 60-kD glycoprotein (gp60) gene [50]. The detection and genotyping of G. duodenalis were based on PCR and sequence analysis of the glutamate dehydrogenase (gdh), beta -giardin (bg) and triosephosphate isomerase (tpi) genes [51], whereas the prevalence and genotypes of E. bieneusi were identified by PCR and sequence analysis of a 392-bp fragment covering the entire internal transcribed spacer (ITS) of the rRNA gene [52]. Two replicates were used in the PCR analysis of each specimen at each locus, with reagent-grade water being used as the negative control. The secondary PCR products were examined by electrophoresis in 1.5% agarose gels and visualized after ethidium bromide staining.
Sequence analysis
All secondary PCR products of the expected size for each genetic locus in agarose gel electrophoresis were sequenced bi-directionally with secondary PCR primers on an ABI 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) at the BioSune Biotechnology Company (Shanghai, China). The sequences obtained were assembled using ChromasPro v.2.1.5.0 (http://technelysium.com.au/ChromasPro.html), edited with BioEdit v.7.1.3.0 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html), and compared with reference sequences from GenBank using ClustalX 2.0.11 (http://clustal.org/) to identify the species, genotypes or subtypes of pathogens under analysis. Nucleotide sequences generated in the study were submitted to the GenBank database under the accession numbers MK252644-MK252647 for Cryptosporidium spp., MK252648-MK252662 and MH893662-MH893688 for G. duodenalis, and MK252663-MK252664 for E. bieneusi.
Statistical analysis
The detection rates of each pathogen were calculated by sampling date, age and clinical sign. The Chi-square test implemented in SPSS Statistics v.20.0 for Windows (IBM Corp., New York, NY, USA) was used to compare the prevalence detection rate of pathogens among sampling dates (spring, summer and winter of northern hemisphere), age groups (weeks), or clinical signs (with or without diarrhea). Differences were considered significant at P < 0.05.
Results
Prevalence of Cryptosporidium spp., G. duodenalis and E. bieneusi
Of the 388 specimens analyzed, 93 (24.0%) were positive for Cryptosporidium spp. There were no significant differences in detection rates among the three batches of sampling conducted during the seasons (Table 1). They ranged from 12.1% (4/33) to 35.7% (5/14) in pre-weaned calves by age in weeks (Table 2).
Based on PCR analysis at any of the three genetic loci, the detection rate of G. duodenalis was 74.2% (288/388). Among the three northern hemisphere seasons, the highest detection rate was 92.3% (166/169) in winter; lower rates were observed in summer (55.8% or 62/111; χ2 = 51.637, df = 1, P < 0.0001) and spring (64.8% or 70/108; χ2 = 33.155, df = 1, P < 0.0001) (Table 1). By age, detection rates increased rapidly from calves of one week in age to calves of nine weeks in age (Table 2). The detection rate was over 60% when the pre-weaned calves were three weeks old. The difference in overall detection rates between 1–2 week-old calves and older calves was significant (χ2 = 32.835, df = 1, P < 0.0001).
The overall detection rate of E. bieneusi was 15.7% (61/388). Enterocytozoon bieneusi detection appeared at older than 2-week-old calves. There was a significant difference in E. bieneusi detection rates between spring (38.0% or 41/108) and summer (7.2% or 8/111) (χ2 = 29.813, df = 1, P < 0.0001) or winter (7.1% or 12/169) (χ2 = 40.563, df = 1, P < 0.0001) (Table 1).
Cryptosporidium spp., G. duodenalis and E. bieneusi detection and presence of diarrhea
The specimens in this study were from calves with (n = 21) and without diarrhea (n = 367). The difference in overall Cryptosporidium detection between the two groups of animals was not significant (χ2 = 0.817, df = 1, P = 0.24). Similarly, the difference in G. duodenalis (χ2 = 0.091, df = 1, P = 0.76) or E. bieneusi (χ2 = 0.526, df = 1, P = 0.23) detection between animals with and without diarrhea was not significant (Table 3).
Distribution of Cryptosporidium species and C. parvum subtype
Based on sequence analysis of the SSU rRNA gene, three Cryptosporidium species were observed in pre-weaned dairy calves, including C. bovis (MK252645, n = 73), C. parvum (MK252646, n = 12) and C. ryanae (MK252644, n = 7) (Table 2). Concurrence of C. bovis and C. parvum was identified in an 8-week-old calf. C. parvum was the dominant species during the first two weeks of life, being detected in five of the six Cryptosporidium-positive calves. The appearance of C. parvum decreased gradually afterwards. In contrast, C. bovis was the dominant species at 3–9 weeks of age, reaching peak occurrence in 6-week-old calves. Similarly, C. ryanae was detected in 4–8-week-old calves. There was no significant association between the occurrence of diarrhea and C. parvum (χ2 = 0.363, df = 1, P = 0.55) or C. bovis (χ2 = 1.215, df = 1, P = 0.27) detection (Table 3). None of the calves with diarrhea were positive for C. ryanae. By sequence analysis of the gp60 gene, 10 of the 12 C. parvum-positive specimens were successfully subtyped as IIdA19G1 (MK252647).
Genotypes and subtypes of G. duodenalis
Of the 288 G. duodenalis-positive specimens, 279 were PCR positive using bg PCR, 209 using tpi and 233 using gdh. Two genotypes were detected among the 288 genotyped specimens, including assemblages E (n = 282) and A (n = 1); five specimens presented both assemblages (Table 1).
Sequence analysis at each genetic locus revealed a high genetic heterogeneity within assemblage E. At the bg locus, 15 subtypes of assemblage E were observed, including six subtypes with sequences identical to those in GenBank: E1 (MK252651, n = 4), E2 (MK252652, n = 13), E3 (MK252653, n = 172), E4 (MK252648, n = 16), E5 (MK252649, n = 17) and E8 (MK252650, n = 18). The remaining nine subtypes represented novel sequences in the literature: E18 (MH893675, n = 3), E19 (MH893678, n = 1), E20 (MH893676, n = 3), E21 (MH893677, n = 1), E22 (MH893682, n = 1), E23 (MH893679, n = 1), E24 (MH893680, n = 1), E25 (MH893681, n = 1) and E26 (MH893683, n = 1). In addition, double peaks were observed in the sequence trace files from 26 assemblage E specimens. Similarly, 17 assemblage E subtypes were identified at the tpi locus, including four known subtypes and 13 novel ones. The former known assemblages included E17 (MK252661, n = 118), E34 (MK252659, n = 14), E1 (MK252662, n = 12) and E18 (MK252660, n = 1), whereas the latter included E35 (MH893664, n = 3), E36 (MH893663, n = 3), E37 (MH893662, n = 20), E38 (MH893667, n = 1), E39 (MH893665, n = 1), E40 (MH893666, n = 1), E41 (MH893670, n = 1), E42 (MH893669, n = 4), E43 (MH893673, n = 2), E44 (MH893674, n = 1), E45 (MH893668, n = 2), E46 (MH893671, n = 1) and E47 (MH893672, n = 1). In contrast, 10 subtypes of assemblage E were identified at the gdh locus, including five known subtypes and five novel ones. The former included E1 (MK252654, n = 113), E3 (MK252657, n = 91), E29 (MK252656, n = 2), E37 (MK252655, n = 1) and E41 (MK252658, n = 1), whereas the latter included E35 (MH893685, n = 1), E36 (MH893686, n = 1), E38 (MH893684, n = 1), E39 (MH893687, n = 1) and E40 (MH893688, n = 1). Double peaks were identified from 23 and 20 assemblage E specimens at the tpi and gdh loci, respectively. The only assemblage A isolate from a 5-week-old calf was identified as subtype A1 at all three loci, representing sub-assemblage AI.
Altogether, 149 specimens were successfully subtyped at all three genetic loci, forming 49 assemblage E MLGs and one assemblage A MLG (Table 4). Because of the presence of double peaks, data from 44 specimens were excluded in the MLG data analysis. MLG-E1 and MLG-E2 were the most common MLGs in pre-weaned dairy calves, being found in 46 and 21 animals, respectively. Among sampling dates, the MLG-E1 prevalence increased from spring to winter; it was significantly different between winter and spring (χ2 = 8.6445, df = 1, P = 0.0033) and spring and summer (χ2 = 8.5132, df = 1, P = 0.0035). In contrast, there were no differences in the prevalence of MLG-E2 between spring and summer (χ2 = 0.5775, df = 1, P = 0.45), spring and winter (χ2 = 1.0172, df = 1, P = 0.31) or summer and winter (χ2 = 3.0958, df = 1, P = 0.08). There were 39 MLGs in winter but only 23 in spring and seven in summer. The number of MLGs was significantly different between spring and summer (χ2 = 10.4040, df = 1, P = 0.0012) and winter and summer (χ2 = 13.7240, df = 1, P = 0.0002).
Genotypes of E. bieneusi
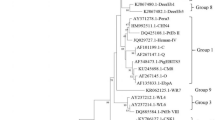
Two known ITS genotypes were detected among the 61 successfully sequenced E. bieneusi specimens, including J (n = 57) and D (n = 3). Between them, J belongs to the host-adapted group 2 while D belongs to the zoonotic group 1. Concurrence of the two genotypes was seen in one animal (Table 1). Among sampling dates, genotype J was detected in all three seasons while genotype D was mostly detected in winter.
Discussion
A high prevalence of Cryptosporidium spp., G. duodenalis and E. bieneusi was found in pre-weaned calves on the study farm in Guangdong. The overall detection rate of Cryptosporidium spp. was 24.0% (93/388). This is similar to detection rates in eastern China (19.7% or 69/350 to 24.3% or 36/148; [8, 53]) and Shaanxi (24.7% or 46/186; [9]), but higher than those reported in northwestern (10.2% or 19/186 to 15.6% or 37/237; [10, 14, 15]), southwestern (14.4% or 40/278; [13]), northern (2.6% or 9/651 to 3.47% or 14/404; [16, 17]), southern (6.4% or 19/297; [43]) and central China (15.8% or 42/265; [12]). Other studies conducted in Shanghai (37.0% or 303/818; [11]), Henan (36.2% or 108/298; [8]), and northeastern China (32.1% or 61/190 to 47.7% or 72/151; [18, 54]) reported higher detection rates of Cryptosporidium spp. In contrast, the 74.2% (288/388) detection rate of G. duodenalis obtained in this study is the highest among detection rates reported in pre-weaned dairy calves in China to date with previous studies reporting detection rates of 2.7% (11/404) to 60.1% (492/818) [12,13,14, 16, 17, 24, 25, 27, 29, 32, 44, 47, 55, 56]. The detection rate of E. bieneusi in this study was found to be 15.7% (61/388), which is within the range of reported detection rates of 2.0% (3/148) to 37.6% (35/93) in China [8, 17, 27, 36, 39,40,41, 46, 57]. Differences in study design, sample size, detection methods, climate, age and management of animals may be another source of variation in reported detection rates from other studies. In the present study, pre-weaned dairy calves were kept in individual stalls and under good management, with a low occurrence of diarrhea (21/388) at the sampling time. Dairy calves with diarrhea were given broad-spectrum antibiotics and oral electrolytes and were separated from healthy calves during the illness.
In the present study, seasonal differences in detection rates were observed in G. duodenalis and E. bieneusi, but not in Cryptosporidium spp. A higher G. duodenalis detection rate was noted in winter, which is consistent with findings in previous studies in Henan Province, China and in the Netherlands [32, 58]. A study of the impact of temperatures on the survival of G. duodenalis observed increased die off of G. duodenalis at higher temperatures [59], which could explain the higher transmission of G. duodenalis in winter in tropical Guangdong. In contrast, the detection rate of E. bieneusi in pre-weaned dairy calves was higher in spring than in summer and winter. This is also in agreement with observations in a previous study conducted in Korea [60]. Differences in the hardiness of environmental stages of Cryptosporidium spp., G. duodenalis, and E. bieneusi could contribute to the differences in seasonal transmission patterns in dairy calves among the three types of pathogens. Prevention and control measures against these enteric pathogens should take these seasonal variations in prevalence into consideration.
Three Cryptosporidium species were observed in pre-weaned dairy calves in Guangdong, including C. bovis, C. parvum and C. ryanae. C. bovis was the dominant species, followed by C. parvum. This result is different from a recent study conducted on ten farms in Guangdong, which observed only C. bovis, C. ryanae and C. andersoni in pre-weaned dairy calves [43]. The presence of zoonotic C. parvum and the absence of C. andersoni in pre-weaned dairy calves in this study are in stern contrast with observations in the previous study in Guangdong. However, C. bovis was the dominant species in both studies, which is in agreement with results in previous studies in China [4, 6]. More studies are needed to better understand the distribution of Cryptosporidium species in Guangdong.
The Cryptosporidium species distribution in pre-weaned dairy calves appears to be different between China and other countries. In a longitudinal study conducted in the USA [61], the detection rate was highest during the first four weeks, largely because of the exclusive presence of C. parvum which has a short patent period. In contrast, C. bovis appears mostly after weaning and can last for weeks or months. Because of the presence of C. bovis, a high overall Cryptosporidium detection rate was recorded throughout early life in dairy calves in a longitudinal study in Shanghai [11]. Similarly, in the present study and a previous study in Henan [7], the detection rates of Cryptosporidium spp. in pre-weaned dairy calves were not significantly different among age groups in weeks. The common presence of both C. parvum and C. bovis in these studies, which had different age-associated occurrence patterns in the present study, is apparently responsible for the persistent detection of Cryptosporidium spp. in pre-weaned dairy calves in China.
The subtype IIdA19G1 identified in the pre-weaned dairy calves in Guangdong is one of the two dominant C. parvum subtypes identified in ruminants, equine animals and rodents in China. To date, five C. parvum IId subtypes have been observed in pre-weaned dairy calves in China, including IIdA14G1, IIdA15G1, IIdA17G1, IIdA19G1 and IIdA20G1, with IIdA15G1 and IIdA19G1 being the common ones [4, 6]. In cattle, subtype IIdA19G1 has been detected in Heilongjiang, Henan, Beijing, Shanghai and Hebei and Tianjin [7, 11, 16, 17, 54] while IIdA15G1 is mostly seen in Ningxia, Sichuan, Gansu, Xinjiang, Beijing and northeastern China [10, 13, 15, 18, 62]. IId subtypes have been found in humans in Europe, Asia and Australia [63,64,65,66,67,68,69,70,71,72,73]. IIdA19G1, which was found in the present study, is a known human pathogen in Portugal, Sweden and China [63, 64, 71]. Both IIdA15G1 and IIdA19G1 are apparently common in various rodents in China [4].
Consistent with previous studies conducted worldwide [12, 17, 24,25,26, 28, 29, 74, 75], G. duodenalis assemblage E was the predominant assemblage identified in pre-weaned dairy calves in Guangdong. In addition, sub-assemblage AI was identified in one animal and concurrence of AI and assemblage E was identified in five animals in this study. As both assemblage E and sub-assemblage AI have been detected in humans in some studies [2, 33,34,35], dairy calves could play a potential role in the transmission of human G. duodenalis infection. Although assemblage E has not thus far been found in humans in China, more studies are needed to monitor its zoonotic transmission.
A very high genetic diversity of assemblage E was found in pre-weaned dairy calves in Guangdong. All three genetic loci in this study showed high sequence polymorphism, yielding 15, 17 and 10 subtypes at the bg, tpi and gdh loci, respectively. The combination of this sequence polymorphism at the three genetic loci leads to the identification of 49 MLGs of assemblage E. A previous study conducted in Shanghai also revealed high genetic diversity of assemblage E in pre-weaned dairy cattle on a single farm [24]. In that study, there were 36 MLGs of assemblage E on Farm 3. The reason for the high subtype diversity of assemblage E remains unclear. Previous studies have suggested that intra-assemblage genetic recombination may be responsible for the high subtype diversity [51, 76]. The high detection rate of G. duodenalis in this study and the existence of multiple subtypes on the farm and double peaks in sequencing trace files from some specimens support the potential role of genetic recombination in generating high genetic heterogeneity within assemblage E [51, 76]. This needs to be substantiated by population genetic studies.
Genotypes J and D were detected in pre-weaned dairy calves in Guangdong, with J as the dominant E. bieneusi genotype. This agrees with the results of most studies in China [17, 36, 39,40,41]. In contrast, genotype I, another common group 2 E. bieneusi genotype in pre-weaned dairy calves worldwide [8, 27, 37, 40,41,42, 77], was not detected in the present study. Genotype J has been found in a few human cases [57]. In contrast genotype D, which belongs to group 1, is a well-known zoonotic genotype [1], and appears to be a common E. bieneusi genotype in humans in China [63, 78, 79]. Thus far, no studies have been conducted to assess the role of zoonotic transmission of genotypes D and J in humans in Guangdong.
Conclusions
Although C. bovis remains as the dominant Cryptosporidium species in pre-weaned dairy calves in Guangdong, there is an increasing presence of C. parvum in these animals, as reported elsewhere in China. The IIdA19G1 subtype of C. parvum identified has been detected recently in humans and other animals in China. In contrast, assemblage E of G. duodenalis and genotype J of E. bieneusi seen in this study are well-known pathogens in dairy cattle in China and elsewhere. As there are increasing numbers of reports of their detection in humans, further molecular epidemiological studies are required to better understand their transmission and public health significance in Guangdong, China.
Abbreviations
- PCR:
-
Polymerase chain reaction
- gp60:
-
60-kDa glycoprotein gene
- MLG:
-
Multilocus genotype
- bg :
-
Beta-giardia
- gdh :
-
Glutamate dehydrogenase
- tpi :
-
Triosephosphate isomerase
- ITS:
-
Internal transcribed spacer
- SSU rRNA:
-
Small subunit rRNA
References
Santín M, Fayer R. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res Vet Sci. 2011;90:363–71.
Feng YY, Xiao LH. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbio Rev. 2011;24:110–40.
Xiao LH. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. 2010;124:80–9.
Feng Y, Xiao L. Molecular epidemiology of cryptosporidiosis in China. Front Microbiol. 2017;8:1701.
Ryan U, Fayer R, Xiao LH. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology. 2014;141:1667–85.
Wang RJ, Zhao GH, Gong YY, Zhang LX. Advances and perspectives on the epidemiology of bovine Cryptosporidium in China in the past 30 years. Front Microbiol. 2017;8:1823.
Wang RJ, Wang HL, Sun YR, Zhang LX, Jian FC, Qi M, et al. Characteristics of Cryptosporidium transmission in preweaned dairy cattle in Henan, China. J Clin Microbiol. 2011;49:1077–82.
Ma J, Li P, Zhao X, Xu H, Wu W, Wang Y, et al. Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Vet Parasitol. 2015;207:220–7.
Qi MZ, Fang YQ, Wang XT, Zhang LX, Wang RJ, Du SZ, et al. Molecular characterization of Cryptosporidium spp. in pre-weaned calves in Shaanxi Province, north-western China. J Med Microbiol. 2015;64:111–6.
Zhang XX, Tan QD, Zhou DH, Ni XT, Liu GX, Yang YC, et al. Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle, northwest China. Parasitol Res. 2015;114:2781–7.
Cai M, Guo YQ, Pan BL, Li N, Wang XL, Tang CX, et al. Longitudinal monitoring of Cryptosporidium species in pre-weaned dairy calves on five farms in Shanghai, China. Vet Parasitol. 2017;241:14–9.
Fan YY, Wang T, Koehler AV, Hu M, Gasser RB. Molecular investigation of Cryptosporidium and Giardia in pre- and post-weaned calves in Hubei Province, China. Parasit Vectors. 2017;10:519.
Zhong ZJ, Dan JM, Yan GW, Tu R, Tian YA, Cao SZ, et al. Occurrence and genotyping of Giardia duodenalis and Cryptosporidium in pre-weaned dairy calves in central Sichuan Province, China. Parasite. 2018;25:45.
Huang J, Yue D, Qi M, Wang R, Zhao J, Li J, et al. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. BMC Vet Res. 2014;10:292.
Qi M, Wang H, Jing B, Wang D, Wang R, Zhang L. Occurrence and molecular identification of Cryptosporidium spp. in dairy calves in Xinjiang, northwestern China. Vet Parasitol. 2015;212:404–7.
Li F, Wang H, Zhang Z, Li J, Wang C, Zhao J, et al. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Beijing, China. Vet Parasitol. 2016;219:61–5.
Hu S, Liu Z, Yan F, Zhang Z, Zhang G, Zhang L, et al. Zoonotic and host-adapted genotypes of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in dairy cattle in Hebei and Tianjin, China. Vet Parasitol. 2017;248:68–73.
Tao W, Li Y, Yang H, Song M, Lu Y, Li W. Widespread occurrence of zoonotic Cryptosporidium species and subtypes in dairy cattle from northeast China: public health concerns. J Parasitol. 2018;104:10–7.
Silverlås C, Näslund K, Björkman C, Mattsson JG. Molecular characterisation of Cryptosporidium isolates from Swedish dairy cattle in relation to age, diarrhoea and region. Vet Parasitol. 2010;169:289–95.
Muhid A, Robertson I, Ng J, Ryan U. Prevalence of and management factors contributing to Cryptosporidium spp. infection in pre-weaned and post-weaned calves in Johor, Malaysia. Exp Parasitol. 2011;127:534–8.
Amer S, Zidan S, Feng YY, Adamu H, Li N, Xiao LH. Identity and public health potential of Cryptosporidium spp. in water buffalo calves in Egypt. Vet Parasitol. 2013;191:123–7.
Becher KA, Robertson ID, Fraser DM, Palmer DG, Thompson RCA. Molecular epidemiology of Giardia and Cryptosporidium infections in dairy calves originating from three sources in Western Australia. Vet Parasitol. 2004;123:1–9.
Azcona-Gutierrez JM, de Lucio A, Hernandez-de-Mingo M, Garcia-Garcia C, Soria-Blanco LM, Morales L, et al. Molecular diversity and frequency of the diarrheagenic enteric protozoan Giardia duodenalis and Cryptosporidium spp. in a hospital setting in northern Spain. PLoS One. 2017;12:e0178575.
Wang XL, Cai M, Jiang W, Wang YP, Jin Y, Li N, et al. High genetic diversity of Giardia duodenalis assemblage E in pre-weaned dairy calves in Shanghai, China, revealed by multilocus genotyping. Parasitol Res. 2017;116:2101–10.
Meng Q, Wang H, Jing B, Wang R, Jian F, Ning C, et al. Prevalence and multilocus genotyping of Giardia duodenalis in dairy calves in Xinjiang, northwestern China. Parasit Vectors. 2016;9:546.
Nguyen ST, Fukuda Y, Nguyen DT, Tada C, Nakai Y. Prevalence and first genotyping of Giardia duodenalis in beef calves in Vietnam. Trop Anim Health Prod. 2016;48:837–41.
Wang XT, Wang RJ, Ren GJ, Yu ZQ, Zhang LX, Zhang SY, et al. Multilocus genotyping of Giardia duodenalis and Enterocytozoon bieneusi in dairy and native beef (Qinchuan) calves in Shaanxi Province, northwestern China. Parasitol Res. 2016;115:1355–61.
Wegayehu T, Karim MR, Erko B, Zhang LX, Tilahun G. Multilocus genotyping of Giardia duodenalis isolates from calves in Oromia Special Zone, central Ethiopia. Infect Genet Evol. 2016;43:281–8.
Jian Y, Zhang X, Li X, Karanis G, Ma L, Karanis P. Prevalence and molecular characterization of Giardia duodenalis in cattle and sheep from the Qinghai-Tibetan Plateau Area (QTPA), northwestern China. Vet Parasitol. 2018;250:40–4.
Abeywardena H, Jex AR, Koehler AV, Rajapakse RP, Udayawarna K, Haydon SR, et al. First molecular characterization of Cryptosporidium and Giardia from bovines (Bos taurus and Bubalus bubalis) in Sri Lanka: unexpected absence of C. parvum from pre-weaned calves. Parasit Vectors. 2014;7:75.
Winkworth CL, Learmonth JJ, Matthaei CD, Townsend CR. Molecular characterization of Giardia isolates from calves and humans in a region in which dairy farming has recently intensified. Appl Environ Microb. 2008;74:5100–5.
Wang HY, Zhao GH, Chen GY, Jian FC, Zhang SM, Feng C, et al. Multilocus genotyping of Giardia duodenalis in dairy cattle in Henan, China. PLoS One. 2014;9:e100453.
Abdel-Moein KA, Saeed H. The zoonotic potential of Giardia intestinalis assemblage E in rural settings. Parasitol Res. 2016;115:3197–202.
Fantinatti M, Bello AR, Fernandes O, Da-Cruz AM. Identification of Giardia lamblia assemblage E in humans points to a new anthropozoonotic cycle. J Infect Dis. 2016;214:1256–9.
Zahedi A, Field D, Ryan U. Molecular typing of Giardia duodenalis in humans in Queensland - first report of Assemblage E. Parasitology. 2017;144:1154–61.
Jiang Y, Tao W, Wan Q, Li Q, Yang Y, Lin Y, et al. Zoonotic and potentially host-adapted Enterocytozoon bieneusi genotypes in sheep and cattle in northeast China and an increasing concern about the zoonotic importance of previously considered ruminant-adapted genotypes. Appl Environ Microbio. 2015;81:3326–35.
Del Coco VF, Córdoba MA, Bilbao G, De ACP, Basualdo JA, Santín M. First report of Enterocytozoon bieneusi from dairy cattle in Argentina. Vet Parasitol. 2014;199:112–5.
Matos O, Lobo ML, Xiao L. Epidemiology of Enterocytozoon bieneusi infection in humans. J Parasitol Res. 2012;2012:981424.
Tang C, Cai M, Wang L, Guo Y, Li N, Feng Y, et al. Genetic diversity within dominant Enterocytozoon bieneusi genotypes in pre-weaned calves. Parasit Vectors. 2018;11:170.
Qi M, Jing B, Jian F, Wang R, Zhang S, Wang H, et al. Dominance of Enterocytozoon bieneusi genotype J in dairy calves in Xinjiang, northwest China. Parasitol Int. 2017;66:960–3.
Li J, Luo N, Wang C, Meng Q, Cao J, Cui Z, et al. Occurrence, molecular characterization and predominant genotypes of Enterocytozoon bieneusi in dairy cattle in Henan and Ningxia, China. Parasit Vectors. 2016;9:142.
Da SFV, Lopes CW, de Oliveira FC, Fayer R, Santin M. New findings of Enterocytozoon bieneusi in beef and dairy cattle in Brazil. Vet Parasitol. 2016;216:46–51.
Liang N, Wu Y, Sun M, Chang Y, Lin X, Yu L, et al. Molecular epidemiology of Cryptosporidium spp. in dairy cattle in Guangdong Province, south China. Parasitology. 2018. https://doi.org/10.1017/S0031182018001129.
Zhang XX, Tan QD, Zhao GH, Ma JG, Zheng WB, Ni XT, et al. Prevalence, risk factors and multilocus genotyping of Giardia intestinalis in dairy cattle, northwest China. J Eukaryot Microbiol. 2016;63:498–504.
Qi M, Wang RJ, Jing B, Jian FC, Ning CS, Zhang LX. Prevalence and multilocus genotyping of Cryptosporidium andersoni in dairy cattle and He cattle in Xinjiang. China. Infect Genet Evol. 2016;44:313–7.
Zhao W, Zhang W, Yang F, Zhang L, Wang R, Cao J, et al. Enterocytozoon bieneusi in dairy cattle in the northeast of China: genetic diversity of ITS gene and evaluation of zoonotic transmission potential. J Eukaryot Microbiol. 2015;62:553–60.
Cui Z, Wang L, Cao L, Sun M, Liang N, Wang H, et al. Genetic characteristics and geographic segregation of Giardia duodenalis in dairy cattle from Guangdong Province, southern China. Infect Genet Evol. 2018;66:95–100.
Jiang JL, Alderisio KA, Singh A, Xiao LH. Development of procedures for direct extraction of Cryptosporidium DNA from water concentrates and for relief of PCR inhibitors. Appl Environ Microb. 2005;71:1135–41.
Ryan U, Xiao LH, Read C, Zhou L, Lal AA, Pavlasek I. Identification of novel Cryptosporidium genotypes from the Czech Republic. Appl Environ Microb. 2003;69:4302–7.
Feng YY, Li N, Duan LP, Xiao LH. Cryptosporidium genotype and subtype distribution in raw wastewater in Shanghai, China: evidence for possible unique Cryptosporidium hominis transmission. J Clin Microbiol. 2009;47:153–7.
Caccio SM, Beck R, Lalle M, Marinculic A, Pozio E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int J Parasitol. 2008;38:1523–31.
Sulaiman IM, Fayer R, Lal AA, Trout JM, Schaefer FW, Xiao L. Molecular characterization of microsporidia indicates that wild mammals harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl Environ Microbiol. 2003;69:4495.
Chen F, Huang KH. Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle from farms in China. J Vet Sci. 2012;13:15–22.
Zhang W, Wang R, Yang F, Zhang L, Cao J, Zhang X, et al. Distribution and genetic characterizations of Cryptosporidium spp. in pre-weaned dairy calves in northeastern China’s Heilongjiang Province. PLoS One. 2013;8:e54857.
Liu G, Su Y, Zhou M, Zhao J, Zhang T, Ahmad W, et al. Prevalence and molecular characterization of Giardia duodenalis isolates from dairy cattle in northeast China. Exp Parasitology. 2015:15420–4.
Liu A, Zhang X, Zhang L, Wang R, Li X, Shu J, et al. Occurrence of bovine giardiasis and endemic genetic characterization of Giardia duodenalis isolates in Heilongjiang Province, in the northeast of China. Parasitol Res. 2012;111:655–61.
Zhang X, Wang Z, Su Y, Liang X, Sun X, Peng S, et al. Identification and genotyping of Enterocytozoon bieneusi in China. J Clin Microbiol. 2011;49:2006–8.
Huetink RE, van der Giessen JW, Noordhuizen JP, Ploeger HW. Epidemiology of Cryptosporidium spp. and Giardia duodenalis on a dairy farm. Vet Parasitol. 2001;102:53–67.
Alum A, Absar IM, Asaad H, Rubino JR, Ijaz MK. Impact of environmental conditions on the survival of Cryptosporidium and Giardia on environmental surfaces. Interdiscip Perspect Infect Dis. 2014;2014:210385.
Lee JH. Prevalence and molecular characteristics of Enterocytozoon bieneusi in cattle in Korea. Parasitol Res. 2007;101:391–6.
Santín M, Trout JM, Fayer R. A longitudinal study of cryptosporidiosis in dairy cattle from birth to 2 years of age. Vet Parasitol. 2008;155:15–23.
Cui Z, Wang R, Huang J, Wang H, Zhao J, Luo N, et al. Cryptosporidiosis caused by Cryptosporidium parvum subtype IIdA15G1 at a dairy farm in northwestern China. Parasit Vectors. 2014;7:529.
Wang L, Zhang HW, Zhao XD, Zhang LX, Zhang GQ, Guo MJ, et al. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J Clin Microbiol. 2013;51:557–63.
Insulander M, Silverlas C, Lebbad M, Karlsson L, Mattsson JG, Svenungsson B. Molecular epidemiology and clinical manifestations of human cryptosporidiosis in Sweden. Epidemiol Infect. 2013;141:1009–20.
Iqbal A, Lim YA, Surin J, Sim BL. High diversity of Cryptosporidium subgenotypes identified in Malaysian HIV/AIDS individuals targeting gp60 gene. PLoS One. 2012;7:e31139.
Chalmers RM, Smith RP, Hadfield SJ, Elwin K, Giles M. Zoonotic linkage and variation in Cryptosporidium parvum from patients in the United Kingdom. Parasitol Res. 2011;108:1321–5.
Hijjawi N, Ng J, Yang RC, Atoum MFM, Ryan U. Identification of rare and novel Cryptosporidium gp60 subtypes in human isolates from Jordan. Exp Parasitol. 2010;125:161–4.
Zintl A, Proctor AF, Read C, Dewaal T, Shanaghy N, Fanning S, et al. The prevalence of Cryptosporidium species and subtypes in human faecal samples in Ireland. Epidemiol Infect. 2009;137:270–7.
Waldron LS, Ferrari BC, Power ML. Glycoprotein 60 diversity in C. hominis and C. parvum causing human cryptosporidiosis in NSW, Australia. Exp Parasitol. 2009;122:124–7.
Wielinga PR, de Vries A, van der Goot TH, Mank T, Mars MH, Kortbeek LM, et al. Molecular epidemiology of Cryptosporidium in humans and cattle in The Netherlands. Int J Parasitol. 2008;38:809–17.
Alves M, Xiao LH, Antunes F, Matos O. Distribution of Cryptosporidium subtypes in humans and domestic and wild ruminants in Portugal. Parasitol Res. 2006;99:287–92.
Abe N, Matsubayashi M, Kimata I, Iseki M. Subgenotype analysis of Cryptosporidium parvum isolates from humans and animals in Japan using the 60-kDa glycoprotein gene sequences. Parasitol Res. 2006;99:303–5.
Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, et al. Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol. 2005;43:2805–9.
Asher AJ, Hose G, Power ML. Giardiasis in NSW: identification of Giardia duodenalis assemblages contributing to human and cattle cases, and an epidemiological assessment of sporadic human giardiasis. Infect Genet Evol. 2016;44:157–61.
Inpankaew T, Jiyipong T, Thadtapong N, Kengradomkij C, Pinyopanuwat N, Chimnoi W, et al. Prevalence and genotype of Giardia duodenalis in dairy cattle from northern and northeastern part of Thailand. Acta Parasitol. 2015;60:459–61.
Aguiar JM, Silva SO, Santos VA, Taniwaki SA, Oliveira TM, Ferreira HL, et al. Evidence of heterozygosity and recombinant alleles in single cysts of Giardia duodenalis. Rev Bras Parasitol Vet. 2016;25:187–95.
Santin M, Fayer R. A longitudinal study of Enterocytozoon bieneusi in dairy cattle. Parasitol Res. 2009;105:141–4.
Wang L, Xiao LH, Duan LP, Ye JB, Guo YQ, Guo MJ, et al. Concurrent infections of Giardia duodenalis, Enterocytozoon bieneusi, and Clostridium difficile in children during a cryptosporidiosis outbreak in a pediatric hospital in China. PLoS Negl Trop Dis. 2013;7:e2437.
Wang T, Fan YY, Koehler AV, Ma GX, Li T, Hu M, et al. First survey of Cryptosporidium, Giardia and Enterocytozoon in diarrhoeic children from Wuhan, China. Infect Genet Evol. 2017;51:127–31.
Acknowledgements
We thank the farm owners and staff for their assistance in sample collection during this study.
Funding
This work was supported by the National Natural Science Foundation of China (grant numbers 31820103014 and 31630078).
Availability of data and materials
The data supporting the conclusions of this article are included within the article. Representative nucleotide sequences generated in this study were submitted to the GenBank database under the accession numbers MK252644-MK252647 for Cryptosporidium spp., MK252648-MK252662 and MH893662-MH893688 for G. duodenalis, and MK252663-MK252664 for E. bieneusi.
Author information
Authors and Affiliations
Contributions
Ya F and LX conceived and designed the experiments; Yu F, XG, KZ and ZY performed the experiments; Yu F, NL, YG, YW and MK analyzed the data; Yu F, Ya F and LX wrote the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
Permission was obtained from the manager of the study dairy farm before collecting specimens. The research protocol was reviewed and approved by the Ethics Committee of East China University of Science and Technology. The dairy calves were handled in accordance with the Animal Ethics Procedure and Guidelines of the People’s Republic of China.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Feng, Y., Gong, X., Zhu, K. et al. Prevalence and genotypic identification of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in pre-weaned dairy calves in Guangdong, China. Parasites Vectors 12, 41 (2019). https://doi.org/10.1186/s13071-019-3310-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-019-3310-5