Abstract
Background
Cryptosporidium is a key genus of parasitic protists that infect humans and other vertebrates (mammals and birds). Birds are typically infected with C. avium, C. baileyi, C. galli and/or C. meleagridis, the latter of which is recognised as being zoonotic. Stimulated by the previous finding of C. meleagridis subtypes IIIbA21G1R1, IIIbA22G1R1 and IIIbA26G1R1 in diarrhoeic children in Wuhan city and environs in Hubei Province, China, we performed a molecular epidemiological survey to explore whether these or similar subtypes might occur in farmed chickens in this province.
Methods
PCR-coupled sequencing analyses of regions in the small subunit (SSU) of the nuclear ribosomal RNA and 60 kDa glycoprotein (gp60) genes were utilised to characterise Cryptosporidium in faecal samples from chickens (n = 471) from 14 farms from six distinct regions in Hubei Province.
Results
Cryptosporidium baileyi (33/471; 7.0%) and C. meleagridis (15/471; 3.2%) were identified in chickens on eight farms in five of the six distinct geographical regions. No significant age-associated difference in the prevalence of C. baileyi was evident, whereas the prevalence of C. meleagridis was significantly higher in younger (≤ 4 months) than in older chickens (> 4 months). For C. meleagridis, two subtype families, IIIb and IIIe, were defined; some of the subtypes (i.e. IIIbA26G1R1b and IIIbA22G1R1c) characterised here matched those identified previously in diarrhoeic children in Wuhan.
Conclusions
This is the first molecular study reporting the genetic identity and prevalence of C. baileyi and C. meleagridis in chickens in Hubei. The findings suggest that C. meleagridis subtypes IIIbA26G1R1b and IIIbA22G1R1c are cross-transmissible between chickens and humans, raising awareness about the significance of birds as potential reservoirs of zoonotic variants of Cryptosporidium. Future studies might focus on investigating the prevalence of ‘zoonotic’ subtypes of Cryptosporidium meleagridis in various species of wild and domesticated birds, and on comparing them with those found in humans in China and other countries.
Similar content being viewed by others
Background
Cryptosporidium is a socioeconomically significant genus of parasitic protists that infect humans and other vertebrates worldwide. Species within this genus are transmitted via the faecal-oral route, often through direct contact with infected people or animals, food and/or water [1,2,3,4], resulting in gastrointestinal disease in mammals, or respiratory and gastrointestinal infections in birds. Clinical signs linked to human cryptosporidiosis include diarrhoea, dehydration, vomiting, wasting and/or weight loss [5, 6], although subclinical infections can occur [7, 8]. Cryptosporidiosis can resolve as immunity develops to clear an infection [9], but chronic disease can develop in at-risk individuals, including young children and people with immuno-suppression or deficiency [10]. In the absence of readily accessible, effective chemotherapeutics and immunoprophylactics [11, 12], chronic cryptosporidiosis can cause death, particularly in patients seriously affected by HIV/AIDS [13, 14].
Molecular epidemiological investigations utilising PCR-coupled sequencing of particular genetic markers, such as those in the small subunit (SSU) of nuclear ribosomal RNA and the 60 kDa glycoprotein (gp60) genes, have shown that human cryptosporidiosis is predominantly caused by Cryptosporidium hominis or C. parvum infection [7, 15], although symptomatic or non-symptomatic infections have been linked to species (n ≥ 17) including C. felis, C. canis and C. meleagridis or various genotypes (n ≥ 4) [15,16,17]. Cryptosporidiosis cases have been associated with human-to-human (anthroponotic) transmission for C. hominis and C. parvum, and animal-to-human (zoonotic) transmission for taxa including C. parvum, C. meleagridis, C. canis and C. felis [7, 15, 16, 18,19,20,21,22].
In a recent molecular epidemiological survey [23], we were surprised to identify C. meleagridis subtype IIIb (specifically IIIbA21G1R1, IIIbA22G1R1 and IIIbA26G1R1) in 2% of 500 children with a history of diarrhoea in Wuhan and environs in Hubei Province, China, although this prevalence was similar to some previous studies of children in other parts of China [24, 25]. Cryptosporidium meleagridis is primarily a pathogen of birds (e.g. chickens, cockatiels, parrots, pigeons and turkeys) [4, 16, 26,27,28,29,30], and this species has been recorded mainly in immuno-compromised persons and in children [11, 31,32,33], with a potential to lead to chronic cryptosporidiosis [34]. The source of C. meleagridis infection in humans was unclear in most published reports, and the assumption has been that birds can be significant reservoirs of this protist, although anthroponotic transmission might also occur [15,16,17, 30]. A study by Chappell et al. [34] established that healthy adults could be infected by C. meleagridis oocysts, presenting with gastrointestinal symptoms including diarrhoea. Based on the findings of our recent investigation [23], we emphasised the need to explore the presence and prevalence of C. meleagridis in domestic and wild birds. In the present study, we take a first step by investigating Cryptosporidium in intensively farmed chickens in Hubei Province using molecular tools.
Methods
Between July and November 2017, 471 fresh faecal samples were collected from chickens of different age groups from 14 medium- to large-sized farms (each with 2000–25,000 broilers or layers on average) in six distinct geographical regions (Huanggang, Suizhou, Wuhan, Xiantao, Xiangyang and Yichang) in Hubei Province, China (Fig. 1; Table 1). For broilers, 14 to 60 samples were collected per farm (with each sample representing 4–5 faecal deposits randomly collected from flocks of 30–100 chickens each). For layers, 19 to 60 samples were collected per farm (with each sample representing a cage of 5–7 chickens). Genomic DNAs were extracted from individual faecal samples using the PowerSoil DNA isolation kit (MoBio, Carlsbad, USA) and frozen at -20 °C.
Individual DNAs were subjected to nested PCR-based amplification and sequencing of regions of the small subunit of the nuclear ribosomal RNA gene (designated pSSU; ~240 bp) [35] and the 60 kDa glycoprotein gene (designated pgp60; ~900 bp) of Cryptosporidium for classification to the genotype and subtype levels [33]. PCR was conducted in 50 μl containing 50 mM KCl and 10 mM Tris-HCl (pH 8.4; Promega, Madison, USA), 3.0 mM of MgCl2, 200 μM of each deoxynucleotide triphosphate, 50 pmol of each primer and 1 U of MangoTaq DNA polymerase (Bioline, London, UK). Known test-positive, test-negative and no-template (including ‘carry-over’) controls were included in each step of each set of PCRs. PCR products were resolved on 1.5% agarose gels, stained with ethidium bromide prior to sequencing. Then, aliquots (5 μl) of individual amplicons (undigested) were treated with the enzyme Exo I and a thermosensitive alkaline phosphatase (FastAP, Thermo Fisher, Carlsbad, USA), according to the manufacturer's instructions, and subjected to automated sequencing (BigDye Terminator v.3.1 chemistry, Applied Biosystems, Foster City, USA) employing the same primers (separately) as used in PCR.
Sequences were aligned using the program MAFFT [36], and alignments manually adjusted employing the program Mesquite v.3.10 [37]. Sequences were then compared with sequence data available via GenBank (NCBI) using BLASTn (Additional file 1: Table S1). Phylogenetic analysis of pSSU or pgp60 sequence data (including selected reference sequences; Additional file 1: Table S1) was conducted by Bayesian inference (BI) using Monte Carlo Markov Chain (MCMC) analysis in MrBayes v.3.2.6 [38]. The likelihood parameters set for BI analysis of pgp60 data were based on the Akaike Information Criteria test [39] in jModeltest v.2.1.7. The number of substitutions (Nst) was set at 6, with an invariant gamma-distribution. Posterior probability (pp) values were calculated by running 2,000,000 generations with four simultaneous tree-building chains. Trees were saved every 100th generation. At the end of each run, the standard deviation of split frequencies was < 0.01, and the potential scale reduction factor approached one. A 50% majority rule consensus tree for each analysis was constructed based on the final 75% of trees generated by BI. Analyses were run three times to ensure convergence and insensitivity to priors. The outgroups used in the phylogenetic analyses of pSSU and pgp60 sequence data sets were C. molnari (GenBank: HM243547) and C. meleagridis subtype IIId (GenBank: DQ067570.1), respectively. The Chi-square test was performed using SPSS Statistics 24 software (IBM, New York, USA).
Results
All 471 individual faecal samples from chickens were analysed molecularly for the presence of Cryptosporidium species, genotypes and subtypes. The pSSU amplicons were generated from 48 of the 471 DNA samples, equating to an overall prevalence of Cryptosporidium of 10.2% (Table 1), with prevalence values ranging from 5.0% to 18.3% on eight of 14 farms from the five of the six geographical regions (Table 1). Cryptosporidium was detected exclusively in layer chickens, but not in broilers (Table 1).
Cryptosporidium was detected in both age groups (Table 2), and young chickens (≤ 4 months) tended to have a higher infection rate (15.1%) than the chickens of > 4 months (11.7%). Cryptosporidium baileyi and C. meleagridis were detected in birds of both age groups. The prevalence of C. baileyi seemed higher in older chickens (> 4 months), although there was no statistical difference between age groups (χ2 = 0.75, df = 1, P = 0.387). In contrast, the prevalence of C. meleagridis was higher in younger chickens (≤ 4 months) (χ2 = 7.8, df = 1, P = 0.005) (Table 2).
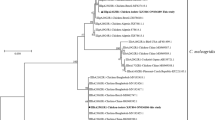
The identification of Cryptosporidium species and genotypes was achieved through the sequencing of pSSU amplicons (n = 48). This analysis revealed C. baileyi in 68.8% and C. meleagridis in 31.2% of the 48 samples; no mixed-species infections were detected. Nine distinct pSSU sequences that represented all 48 samples and both Cryptosporidium species were deposited under GenBank accession numbers MG969393-MG969401, and the relationships of these sequences with selected reference sequences from GenBank (Additional file 1: Table S1) were established through a phylogenetic analysis (Fig. 2). Specifically, eight of these sequences (GenBank: MG969393-MG969400) clustered with known pSSU reference sequences for C. baileyi, and the other one (GenBank: MG969401) clustered with a representative sequence for C. meleagridis, on a branch with ‘zoonotic’ species including C. felis, C. hominis and C. parvum (see Fig. 2).
Relationships among Cryptosporidium taxa inferred from the phylogenetic analysis of sequences from a portion of the small subunit of the nuclear ribosomal RNA gene (pSSU) by Bayesian inference (BI). Posterior probabilities of > 0.95 are indicated at all major nodes. Bold-type indicates Cryptosporidium species or genotypes characterised from faecal DNA samples in this study. The GenBank accession number precedes the species designation; the number of samples of a particular species/genotype is indicated in parentheses. The scale-bar represents the number of substitutions per site. Cryptosporidium molnari was used as an outgroup
Subtyping was achieved through an analysis of sequence data derived from gp60 amplicons obtained (n = 13; deposited under GenBank: MG969387-MG969392). Sequence alignment and phylogenetic analysis of the sequences revealed six new subtypes of C. meleagridis in chickens. This analysis defined two subtype families (IIIb and IIIe; Fig. 3); the commonest subtype family IIIb [IIIbA22G1R1c (n = 6); IIIbA23G1R1d (n = 2) and IIIbA26G1R1b (n = 1)] was identified for nine samples, and subtype family IIIe [IIIeA17G2R1 (n = 1), IIIeA19G2R1 (n = 1) and IIIeA26G2R1 (n = 2)] for four samples. The phylogenetic analysis showed that nine sequences representing C. meleagridis subtype IIIb (GenBank: MG969390-MG969392) clustered with pgp60 sequences (GenBank: KY575457-KY575459) derived from samples from diarrhoeic children from Wuhan [23]. The pgp60 gene sequences representing C. meleagridis subtype IIIb from chickens (GenBank: MG969390-MG969392) showed high sequence similarity (92.7–100%) with those from humans (GenBank: KY575457-KY575459), being associated with subtypes IIIbA22G1R1c, IIIbA23G1R1d and IIIbA26G1R1b.
Relationships among Cryptosporidium meleagridis subtypes inferred from the phylogenetic analysis of sequences from a portion of the nuclear 60-kDa glycoprotein gene (pgp60) by Bayesian inference (BI). Posterior probabilities of > 0.95 are indicated at all major nodes. Bold-type indicates subtypes characterised from faecal DNA samples tested in this study. The GenBank accession number precedes the species designation; the number of samples of a particular species/genotype is indicated in parentheses. The scale-bar represents the number of substitutions per site. Subtype IIIhA7 was used as an outgroup
Discussion
The present molecular investigation reports, for the first time, the presence and prevalence of Cryptosporidium in chickens on commercial farms in Hubei Province, although previous studies have recorded Cryptosporidium in chickens in Henan [40] and Zhejiang [41]. Here, both C. baileyi and C. meleagridis were identified using PCR-based tools. The overall prevalence of Cryptosporidium in chickens was ~10%, which is consistent with percentages recorded previously in China (8.9%, Henan; 9.9%, Zhejiang) [40, 41] and Syria (9.9%) [42], higher than Jordan (4.8%) [43] and Tunisia (4.5%) [44], and lower than Brazil (12.6%) [45]. Cryptosporidium infection has been recorded mainly in broiler chickens in countries including China, Algeria, Germany, Iran, Syria and Tunisia [26, 41, 42, 44, 46, 47]. Published studies indicate that young birds are more frequently infected with Cryptosporidium than adults [28, 40, 46], but more work is needed to confirm such an age-related association.
Cryptosporidium baileyi was detected in most (69%) of the 48 test-positive chicken faecal samples. Cryptosporidium baileyi was originally isolated from commercial broiler chickens [48], has been recorded in a broad range of avian hosts and is considered to be a dominant species in birds, although other taxa including C. avium, C. galli and/or C. meleagridis occur, of which only the latter species is recognised as zoonotic [4, 30, 49]. In China, C. baileyi has been reported in farmed and wild birds, including chickens, quails, ostriches, Pekin ducks, domestic pigeons as well as some pet birds (e.g. rufous turtle dove, zebra finch, red-billed leiothrix, black-billed magpie and white Java sparrow) [27, 40, 50,51,52,53,54]. Previous reports indicate that C. baileyi causes reduced weight gain in broilers and decreased egg production in layer chickens, often in the absence of obvious clinical signs [55, 56]. We propose that C. baileyi may be a species of economic and/or clinical importance, given its relatively broad distribution on most farms studied here, with the exception of Yichang. Although C. baileyi has been recorded in an immunodeficient human patient [57], the zoonotic potential of this species is questionable.
Cryptosporidium meleagridis was detected in almost one third (31%) of the 48 test-positive chicken faecal samples. Although commonly identified in avian hosts, C. meleagridis has been found in humans in a number of countries, including Australia, South Africa, China, France, India, Indonesia, Japan, Jordan, Kenya, Nigeria, Peru, Poland, Portugal, Spain, Sweden, Thailand, Tunisia, the United Kingdom and Uzbekistan [23, 31, 33, 58,59,60,61,62,63,64,65,66,67,68,69,70]. Both immunocompetent and immunocompromised humans can be infected/affected, indicating the public health significance of this species. In China, C. meleagridis has been recorded previously in paediatric patients, HIV-positive individuals in Shanghai and Henan provinces, respectively, and animal contact has been discussed as a significant risk factor [24, 68]; moreover, there is clear molecular evidence of C. meleagridis subtypes being shared by humans and birds. For instance, in Sweden, an outbreak of C. meleagridis infection/cryptosporidiosis in people was confirmed by PCR-based sequence analyses of SSU and heat shock protein 70 (hsp70) gene regions; C. meleagridis genotype I (GenBank: AF12574) defined in humans was the same as found in chickens [21]. In Peru, two subtypes of C. meleagridis (MLG1 and MLG8) characterised from HIV/AIDS patients were shared by birds (chicken, pigeon or duck) in the same location (Lima, Peru) through genetic analyses of gp60 and mini-satellites [71]. Importantly, pgp60 subtypes of C. meleagridis (IIIbA22G1R1c and IIIbA26G1R1b) characterised from diarrhoeic children in Wuhan [23] match those identified in chickens in the present study. These findings suggest that, in Wuhan and environs, chickens may contribute to the transmission of C. meleagridis to humans. Whether wild or other domesticated (e.g. pet) birds might be involved in such transmission requires detailed investigation.
In the present study of chickens, we defined subtype families IIIb and IIIe of C. meleagridis based on pgp60 sequence data. Both of these families have been recorded previously, mainly in humans and occasionally in birds (cockatiel and turkey) and cattle in countries including China, Indonesia, Japan, Kenya, Peru, Thailand and the USA [33]. Within these families, subtypes IIIbA22G1R1c, IIIbA26G1R1b, IIIeA17G2R1, IIIeA19G2R1 and IIIeA26G2R1 have been recorded in humans [33, 68, 72], and were identified here in chickens in Hubei Province. Subtype IIIbA22G1R1c was the predominant subtype in this and our previous study of children [23]. This subtype has been detected in people with travel-acquired infections. For example, subtype IIIbA22G1R1c, detected in some people from Sweden, was linked to travel to India or Thailand [21, 33]. The same subtype had been recorded in an English patient proposed to have become infected in India (sample analysed in the UK) [33, 69]. This information highlights the potential of this pathogen and various IIIb subtypes to be spread worldwide through human travel. In this context, subtype IIIbA23G1R1d was found here, for the first time, in chickens. However, subtype IIIbA23G1R1 had been recorded in quail in Brazil, and a human in Peru [71, 73], and IIIbA23G1R1a, IIIbA23G1R1b and IIIbA23G1R1c in people with histories of travel to Thailand, Indonesia and Malaysia, respectively [21, 33, 74]. We propose that subtype IIIbA23G1R1d might be transmissible from poultry to humans, but this needs to be explored in detail.
Conclusions
This is the first molecular study reporting the genetic identity and prevalence of C. baileyi and C. meleagridis in chickens in Hubei. The findings suggest that C. meleagridis subtypes IIIbA26G1R1b and IIIbA22G1R1c are cross-transmissible between chickens and humans, raising awareness about the significance of birds as potential reservoirs of zoonotic variants of Cryptosporidium. Taken together, this information emphasises the need for epidemiological studies of Cryptosporidium, particularly C. meleagridis, in humans and in birds using accurate diagnostic and analytical tools utilising multiple informative genetic loci. Such investigations should be focused on assessing the transmission patterns and dynamics of cryptosporidiosis.
Abbreviations
- pSSU :
-
a portion of the small subunit of nuclear ribosomal RNA gene
- pgp60 :
-
a portion of the 60 kDa glycoprotein gene
References
Ryan U, Fayer R, Xiao L. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology. 2014;141:1667–85.
Ryan U, Paparini A, Monis P, Hijjawi N. It’s official - Cryptosporidium is a gregarine: what are the implications for the water industry? Water Res. 2016;105:305–13.
Ryan U, Hijjawi N, Xiao L. Foodborne cryptosporidiosis. Int J Parasitol. 2018;48:1–12.
Nakamura AA, Meireles MV. Cryptosporidium infections in birds - a review. Rev Bras Parasitol Vet. 2015;24:253–67.
Mmbaga BT, Houpt ER. Cryptosporidium and Giardia infections in children: a review. Pediatr Clin North Am. 2017;64:837–50.
Checkley W, White AC Jr, Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect Dis. 2015;15:85–94.
Lim YAL, Jex AR, Smith HV, Gasser RB. Cryptosporidiosis in Southeast Asia: what’s out there? Adv Parasitol. 2010;71:1–31.
Jex AR, Chalmers RM, Smith HV, Widmer G, McDonald V, Gasser RB. Cryptosporidiosis. In: Palmer SR, Soulsby EJL, Torgerson PR, Brown DWG, editors. Oxford Textbook of Zoonoses. New York: Oxford University Press; 2011. p. 536–8.
Riggs MW. Recent advances in cryptosporidiosis: the immune response. Microbes Infect. 2002;4:1067–80.
Chalmers RM, Davies AP, Ryan U, Power M, Xiao LH. Clinical cryptosporidiosis. Exp Parasitol. 2010;124:138–46.
Cama VA, Ross JM, Crawford S, Kawai V, Chavez-Valdez R, Vargas D, et al. Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. J Infect Dis. 2007;196:684–91.
Pozio E, Morales MA. The impact of HIV-protease inhibitors on opportunistic parasites. Trends Parasitol. 2005;21:58–63.
Amadi B, Kelly P, Mwiya M, Mulwazi E, Sianongo S, Changwe F, et al. Intestinal and systemic infection, HIV, and mortality in Zambian children with persistent diarrhea and malnutrition. J Pediatr Gastroenterol Nutr. 2001;32:550–4.
Miao YM, Awad-El-Kariem FM, Franzen C, Ellis DS, Müller A, Counihan HM, et al. Eradication of cryptosporidia and microsporidia following successful antiretroviral therapy. J Acquir Immune Defic Syndr. 2000;25:124–9.
Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. 2010;124:80–9.
Zahedi A, Paparini A, Jian F, Robertson I, Ryan UM. Public health significance of zoonotic Cryptosporidium species in wildlife: critical insights into better drinking water management. Int J Parasitol Parasites Wildl. 2016;5:88–109.
Xiao L, Feng Y. Zoonotic cryptosporidiosis. FEMS Immunol Med Microbiol. 2013;52:309–23.
Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, et al. Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol. 2005;43:2805–9.
Hunter PR, Hadfield SJ, Wilkinson D, Lake IR, Harrison FC, Chalmers RM. Subtypes of Cryptosporidium parvum in humans and disease risk. Emerg Infect Dis. 2007;13:82–8.
Alves M, Xiao L, Antunes F, Matos O. Distribution of Cryptosporidium subtypes in humans and domestic and wild ruminants in Portugal. Parasitol Res. 2006;99:287–92.
Silverlås C, Mattsson JG, Insulander M, Lebbad M. Zoonotic transmission of Cryptosporidium meleagridis on an organic Swedish farm. Int J Parasitol. 2012;42:963–7.
Xiao L, Cama VA, Cabrera L, Ortega Y, Pearson J, Gilman RH. Possible transmission of Cryptosporidium canis among children and a dog in a household. J Clin Microbiol. 2007;45:2014–6.
Wang T, Fan Y, Koehler AV, Ma G, Li T, Hu M, et al. First survey of Cryptosporidium, Giardia and Enterocytozoon in diarrhoeic children from Wuhan, China. Infect Genet Evol. 2017;51:127–31.
Feng Y, Wang L, Duan L, Gomez-Puerta LA, Zhang L, Zhao X, et al. Extended outbreak of cryptosporidiosis in a pediatric hospital, China. Emerg Infect Dis. 2012;18:312–4.
Zhang SX, Zhou YM, Xu W, Tian LG, Chen JX, Chen SH, et al. Impact of co-infections with enteric pathogens on children suffering from acute diarrhea in southwest China. Infect Dis Poverty. 2016;5:64.
Baroudi D, Khelef D, Goucem R, Adjou KT, Adamu H, Zhang H, et al. Common occurrence of zoonotic pathogen Cryptosporidium meleagridis in broiler chickens and turkeys in Algeria. Vet Parasitol. 2013;196:334–40.
Li J, Lin X, Zhang L, Qi N, Liao S, Lv M, et al. Molecular characterization of Cryptosporidium spp. in domestic pigeons (Columba livia domestica) in Guangdong Province, southern China. Parasitol Res. 2015;114:2237–41.
Zhang XX, Zhang NZ, Zhao GH, Zhao Q, Zhu XQ. Prevalence and genotyping of Cryptosporidium infection in pet parrots in North China. Biomed Res Int. 2015;2015:549798.
Abe N, Iseki M. Identification of Cryptosporidium isolates from cockatiels by direct sequencing of the PCR-amplified small subunit ribosomal RNA gene. Parasitol Res. 2004;92:523–6.
Ryan UM. Cryptosporidium in birds, fish and amphibians. Exp Parasitol. 2010;124:113–20.
Cama VA, Bern C, Roberts J, Cabrera L, Sterling CR, Ortega Y, et al. Cryptosporidium species and subtypes and clinical manifestations in children, Peru. Emerg Infect Dis. 2008;14:1567–74.
Morgan UM, Weber R, Xiao L, Sulaiman I, Thompson RC, Ndiritu W, et al. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J Clin Microbiol. 2000;38:1180–3.
Stensvold CR, Beser J, Axén C, Lebbad M. High applicability of a novel method for gp60-based subtyping of Cryptosporidium meleagridis. J Clin Microbiol. 2014;52:2311–9.
Chappell CL, Okhuysen PC, Langer-Curry RC, Akiyoshi DE, Widmer G, Tzipori S. Cryptosporidium meleagridis: infectivity in healthy adult volunteers. Am J Trop Med Hyg. 2011;85:238–42.
Xiao L, Singh A, Limor J, Graczyk TK, Gradus S, Lal A. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl Environ Microbiol. 2001;67:1097–101.
Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66.
Maddison W, Maddison DR. Mesquite: a modular system for evolutionary analysis. Evolution. 2018; Version 3.40; https://mesquiteproject.org/
Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5.
Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772.
Wang R, Jian F, Sun Y, Hu Q, Zhu J, Wang F, et al. Large-scale survey of Cryptosporidium spp. in chickens and Pekin ducks (Anas platyrhynchos) in Henan, China: prevalence and molecular characterization. Avian Pathol. 2010;39:447–51.
Wang L, Xue X, Li J, Zhou Q, Yu Y, Du A. Cryptosporidiosis in broiler chickens in Zhejiang Province, China: molecular characterization of oocysts detected in fecal samples. Parasite. 2014;21:36.
Kassouha M. First detection of Cryptosporidium spp. in broiler chickens in Syria. Basrah J Vet Res. 2014;11:253–9.
Hijjawi N, Mukbel R, Yang R, Ryan U. Genetic characterization of Cryptosporidium in animal and human isolates from Jordan. Vet Parasitol. 2016;228:116–20.
Soltane R, Guyot K, Dei-Cas E, Ayadi A. Prevalence of Cryptosporidium spp. (Eucoccidiorida: Cryptosporiidae) in seven species of farm animals in Tunisia. Parasite. 2007;14:335.
Santana BN, Kurahara B, Nakamura AA, da Silva CV, Ferrari ED, da Silva GS, et al. Detection and characterization of Cryptosporidium species and genotypes in three chicken production systems in Brazil using different molecular diagnosis protocols. Prev Vet Med. 2018;151:73–8.
Helmy YA, Krücken J, Abdelwhab EM, Samson-Himmelstjerna GV, Hafez HM. Molecular diagnosis and characterization of Cryptosporidium spp. in turkeys and chickens in Germany reveals evidence for previously undetected parasite species. PLoS One. 2017;12:e0177150.
Shemshadi B, Bahadori SR, Mozafari A. Study on cryptosporidiosis incidence in broilers in Garmsar region, Iran. Comp Clin Path. 2011;20:143–9.
Current WL, Upton SJ, Haynes TB. The life cycle of Cryptosporidium baileyi n. sp. (Apicomplexa, Cryptosporidiidae) infecting chickens. J Protozool. 1986;33:289–96.
Morgan UM, Monis PT, Xiao L, Limor J, Sulaiman I, Raidal S, et al. Molecular and phylogenetic characterisation of Cryptosporidium from birds. Int J Parasitol. 2001;31:289–96.
Amer S, Wang C, He H. First detection of Cryptosporidium baileyi in Ruddy Shelduck (Tadorna ferruginea) in China. J Vet Med Sci. 2010;72:935–8.
Gu Y, Wang X, Zhou C, Li P, Xu Q, Zhao C, et al. Investigation on Cryptosporidium infections in wild animals in a zoo in Anhui Province. J Zoo Wildl Med. 2016;47:846–54.
Qi M, Huang L, Wang R, Xiao L, Xu L, Li J, et al. Natural infection of Cryptosporidium muris in ostriches (Struthio camelus). Vet Parasitol. 2014;205:518–22.
Qi M, Wang R, Ning C, Li X, Zhang L, Jian F, et al. Cryptosporidium spp. in pet birds: genetic diversity and potential public health significance. Exp Parasitol. 2011;128:336–40.
Wang R, Wang F, Zhao J, Qi M, Ning C, Zhang L, et al. Cryptosporidium spp. in quails (Coturnix coturnix japonica) in Henan, China: molecular characterization and public health significance. Vet Parasitol. 2012;187:534–7.
Goodwin MA, Brown J, Resurreccion RS, Smith JA. Respiratory coccidiosis (Cryptosporidium baileyi) among northern Georgia broilers in one company. Avian Dis. 1996;40:572–5.
Sréter T, Varga I. Cryptosporidiosis in birds - a review. Vet Parasitol. 2000;87:261–79.
Ditrich O, Palkovič L, Štěrba J, Prokopič J, Loudová J, Giboda M. The first finding of Cryptosporidium baileyi in man. Parasitol Res. 1991;77:44–7.
Essid R, Mousli M, Aoun K, Abdelmalek R, Mellouli F, Kanoun F, et al. Identification of Cryptosporidium species infecting humans in Tunisia. Am J Trop Med Hyg. 2008;79:702–5.
Gatei W, Greensill J, Ashford RW, Cuevas LE, Parry CM, Cunliffe NA, et al. Molecular analysis of the 18S rRNA gene of Cryptosporidium parasites from patients with or without human immunodeficiency virus infections living in Kenya, Malawi, Brazil, the United Kingdom, and Vietnam. J Clin Microbiol. 2003;41:1458–62.
Guyot K, Follet-Dumoulin A, Lelièvre E, Sarfati C, Rabodonirina M, Nevez G, et al. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J Clin Microbiol. 2001;39:3472–80.
Hijjawi N, Ng J, Yang R, Atoum MF, Ryan U. Identification of rare and novel Cryptosporidium GP60 subtypes in human isolates from Jordan. Exp Parasitol. 2010;125:161–4.
Matos O, Alves M, Xiao L, Cama V, Antunes F. Cryptosporidium felis and C. meleagridis in persons with HIV, Portugal. Emerg Infect Dis. 2004;10:2256–7.
Molloy SF, Smith HV, Kirwan P, Nichols RA, Asaolu SO, Connelly L, et al. Identification of a high diversity of Cryptosporidium species genotypes and subtypes in a pediatric population in Nigeria. Am J Trop Med Hyg. 2010;82:608–13.
Ng-Hublin JS, Combs B, Mackenzie B, Ryan U. Human cryptosporidiosis diagnosed in Western Australia: a mixed infection with Cryptosporidium meleagridis, the Cryptosporidium mink genotype, and an unknown Cryptosporidium species. J Clin Microbiol. 2013;51:2463–5.
Tiangtip R, Jongwutiwes S. Molecular analysis of Cryptosporidium species isolated from HIV-infected patients in Thailand. Trop Med Int Health. 2010;7:357–64.
Wesołowska M, Szostakowska B, Kicia M, Sak B, Kvac M, Knysz B. Cryptosporidium meleagridis infection: the first report in Poland of its occurrence in an HIV-positive woman. Ann Parasitol. 2016;62:239–41.
Yagita K, Izumiyama S, Tachibana H, Masuda G, Iseki M, Furuya K, et al. Molecular characterization of Cryptosporidium isolates obtained from human and bovine infections in Japan. Parasitol Res. 2001;87:950–5.
Wang L, Zhang H, Zhao X, Zhang L, Zhang G, Guo M, et al. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J Clin Microbiol. 2013;51:557–63.
Abal-Fabeiro JL, Maside X, Bello X, Llovo J, Bartolome C. Multilocus patterns of genetic variation across Cryptosporidium species suggest balancing selection at the gp60 locus. Mol Ecol. 2013;22:4723–32.
Abu Samra N, Thompson PN, Jori F, Frean J, Poonsamy B, du Plessis D, et al. Genetic characterization of Cryptosporidium spp. in diarrhoeic children from four provinces in South Africa. Zoonoses Public Health. 2013;60:154–9.
Wang Y, Yang W, Cama V, Wang L, Cabrera L, Ortega Y, et al. Population genetics of Cryptosporidium meleagridis in humans and birds: evidence for cross-species transmission. Int J Parasitol. 2014;44:515–21.
Ng J, Mackenzie B, Ryan U. Longitudinal multi-locus molecular characterisation of sporadic Australian human clinical cases of cryptosporidiosis from 2005 to 2008. Exp Parasitol. 2010;125:348–56.
da Cunha MJR, Cury MC, Santín M. Molecular characterization of Cryptosporidium spp. in poultry from Brazil. Res Vet Sci. 2018;118:331–5.
Insulander M, Silverlås C, Lebbad M, Karlsson L, Mattsson JG, Svenungsson B. Molecular epidemiology and clinical manifestations of human cryptosporidiosis in Sweden. Epidemiol Infect. 2013;141:1009–20.
Acknowledgements
We thank the farmers for donating faecal samples from chickens.
Funding
This project was supported by Huazhong Agricultural University Scientific & Technological Self Innovation Foundation (Program no. 2015RC005), and by the Australian Research Council, The University of Melbourne and Melbourne Water Corporation (RBG et al.).
Availability of data and materials
The data supporting the conclusions of this article are included within the article and its additional file. Nucleotide sequence data reported in this article are publicly available in the GenBank database under accession numbers MF969387-MG969401.
Author information
Authors and Affiliations
Contributions
TW and RBG planned the study. CL and YF collected samples. CL and TW undertook the laboratory and analytical work. TW, RBG and CL wrote the manuscript, with active inputs from AVK and MH. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Table S1. Summary of information on the reference sequences from the GenBank database used in the present study. (XLSX 21 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liao, C., Wang, T., Koehler, A.V. et al. Molecular investigation of Cryptosporidium in farmed chickens in Hubei Province, China, identifies ‘zoonotic’ subtypes of C. meleagridis. Parasites Vectors 11, 484 (2018). https://doi.org/10.1186/s13071-018-3056-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-018-3056-5