Abstract
Background
Spirocerca lupi, the dog oesophageal nematode, causes a potentially fatal disease in domestic dogs, and is currently clinically diagnosed by coproscopy and oesophagoscopy. To date, a single molecular method, a semi-nested PCR, targeting the cox1 gene, has been developed to aid in the diagnosis of spirocercosis. The present study describes three novel high-resolution melt (HRM) quantitative PCR (qPCR) assays targeting fragments of the ITS1, 18S and cytb loci of S. lupi. The performance of these molecular assays in feces was compared to fecal flotation and to the previously described cox1 gene semi-nested PCR in 18 fecal samples from dogs with clinical oesophageal spirocercosis diagnosed by oesophagoscopy.
Results
The HRM qPCR for ITS1 and 18S were both able to detect 0.2 S. lupi eggs per gram (epg), while the HRM qPCR for the cytb and the semi-nested PCR for the cox1 detected 6 epg and 526 epg, respectively. Spirocerca lupi was detected in 61.1%, 44.4%, 27.8%, 11.1% and 5.6% of the fecal samples of dogs diagnosed with spirocercosis by using the ITS1 and 18S HRM qPCR assays, fecal flotation, cytb HRM qPCR and cox1 semi-nested PCR, respectively. All dogs positive by fecal flotation were also positive by ITS1 and 18S HRM qPCRs. Quantification of S. lupi eggs was successfully achieved in the HRM qPCRs and compared to the fecal flotation with no significant difference in the calculated concentrations between the HRM qPCRs that detected the 18S and ITS1 loci and the fecal flotation. The HRM qPCR for the 18S cross-amplified DNA from Toxocara canis and Toxascaris leonina. In contrast, the HRM qPCR for ITS1 did not cross-amplify DNA from other canine gastrointestinal parasites.
Conclusions
This study presents two new molecular assays with significantly increased sensitivity for confirming and quantifying fecal S. lupi eggs. Of these, the HRM qPCR for ITS1 showed the best performance in terms of the limit of detection and absence of cross-amplification with other parasites. These assays will be useful in detecting infection and for follow-up during therapy.
Similar content being viewed by others
Background
Spirocerca lupi, the dog oesophageal nematode, is the etiological agent of spirocercosis, a chronic, potentially life-threatening disease of carnivores, predominantly of canids, including the domestic dog [1]. It has an indirect life-cycle which involves dung beetles as intermediate hosts and canids as definitive hosts. In the dog, S. lupi adults typically localize within oesophageal nodules and shed eggs to the host’s gastrointestinal tract and in the feces [1]. Spirocercosis has been reported in the tropical and subtropical areas of Europe, Africa, Asia and the Americas, with incidences ranging from 5.4% in South Africa [2] to 25% in Iran [3]. The annual incidence in Israel ranges from 0.7 to 8% [4].
Spirocercosis in dogs is suspected based on the history, clinical signs and imaging findings (i.e. thoracic survey radiography and computed tomography), and diagnosis is currently confirmed by coproscopy, using several fecal flotation methods and oesophagoscopy. However, due to the chronicity of this disease, dogs are usually diagnosed in advanced disease stages, which are associated with severe detectable clinical signs [5]. Moreover, clinical signs of spirocercosis are variable, depending on the migratory routes of the worm, occurrence of the transformation of benign to malignant nodules and the parasite intensity. The serological cross-reactivity between S. lupi and Dirofilaria immitis circulating antigen [6] adds to the challenges in the correct diagnosis of spirocercosis [1].
Three laboratory-based diagnostic tools have been described to date for detecting S. lupi infection. These methods include microscopic examination of feces for the identification of S. lupi eggs using flotation methods, such as a technique based on a concentrated sugar solution [7], an immunofluorescence antibody test for detecting S. lupi antigens, which is commercially unavailable, and its utility in clinical practice is unknown [8], and a semi-nested PCR, targeting a 400 bp fragment of the mitochondrial cox1 gene [5]. These methods, however, have limitations. The fecal flotation method requires expertise in distinguishing S. lupi eggs from eggs of other helminths and may produce false negatives due to intermittent egg shedding in the feces [1], while the semi-nested PCR is susceptible to contamination [9, 10]. Thus, sensitive and specific diagnostic tools are needed for early detection of S. lupi in infected dogs.
Quantitative real-time PCR (qPCR) assays provide a very sensitive tool for diagnosing and quantifying parasitic infections [11]. In addition, qPCR, followed by high-resolution melt analysis (HRM), is a useful tool for the specific identification of DNA amplicons based on their melting temperatures, thereby facilitating rapid diagnosis [11]. The present study explored the use of HRM qPCR assays for the specific diagnosis and quantification of S. lupi in dogs with naturally occurring spirocercosis. Accordingly, three different HRM qPCRs, targeting fragments of the 18S ribosomal RNA gene, the internal transcribed spacer 1 (ITS1) loci and cytochrome b gene (cytb) of S. lupi, were developed. In addition, their performance was compared with the currently available diagnostic methods, namely coproscopy by fecal flotation and the cox1 semi-nested PCR. Lastly, the potential cross-amplification of other canine parasitic nematode DNA was evaluated for all the HRM qPCR assays.
Methods
Spirocerca lupi infected dogs and fecal flotation
Fecal samples were collected from 18 dogs (11 males and 7 females), with ages ranging between 1 and 13 years, untreated previously with anthelminthic drugs, and diagnosed with clinical spirocercosis. Oesophageal spirocercosis was suspected based on compatible history and clinical signs and confirmed by characteristic oesophagoscopy findings in all dogs. The latter included identifying oesophageal nodules with a smooth appearance, containing a characteristic nipple-like protuberance [1]. Dogs with neoplastic transformation were excluded [1]. Then, the dogs were treated, and the diagnosis was further confirmed by repeated oesophagoscopy, at weeks 16 to 20 after the initial oesophagoscopy, during treatment to monitor the response to therapy. Fecal samples were examined by a double centrifugation fecal flotation, using sugar solution with a specific gravity of 1.27, as previously described [7], and the number of eggs per gram (epg) of feces was determined in all the positive fecal samples. Approximately 200 mg of each fecal sample were placed in a microcentrifuge tube, and stored at -80 °C pending further analyses.
Collection of eggs from adult S. lupi-female worms
Spirocerca lupi eggs were obtained from female adults maintained in an in vitro culture, as previously described [12]. The growth medium was collected after 24 h of incubation, and the presence of shed eggs contained in the solution was confirmed by microscopic examination. The eggs were maintained at 4 °C pending further analyses. The concentration of the egg suspension was adjusted to 14,200 epg by dilution with the growth medium solution.
DNA extraction of S. lupi samples
DNA was extracted from 200 μl of the in vitro obtained S. lupi-egg suspensions and from 200 mg of the dogs’ fecal samples using the QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions, with the following modifications. Briefly, the weight of each fecal sample used for the extraction was recorded to estimate the egg concentration by the molecular methods. Suspension with the InhibitEX buffer (Qiagen) was heated at 95 °C for 15 min with light mixing by vortex every 5 min. After incubation, five sterile glass beads were added to each sample and mixed by vortex for 10 min. The suspension was then centrifuged at 14,000× rpm for 1 min, and 200 μl of the supernatant was incubated with AL buffer (Qiagen) and proteinase K (Qiagen) for 20 min, at 70 °C, after which, all steps as recommended by the manufacturer were followed. DNA purity and concentration was verified using a NanoDrop spectrophotometer (Thermo-Fisher Scientific, Massachusetts, USA).
Spirocerca lupi standard curve
DNA from the suspension with 14,200 epg of S. lupi was serially diluted 3-fold, to a final volume of 100 μl in PCR-grade water (Biological Industries, Kibbutz Beit-Haemek, Israel). In total, 12 S. lupi-standard concentrations, ranging from 14,200 to 0.07 epg, were prepared for the qPCR analyses. The egg standards were run in triplicates to build a standard curve, and estimate the detection limit, slope, intercept, linear regression coefficient and efficiency for each of the HRM qPCRs.
HRM qPCR assays for the detection of S. lupi DNA
HRM qPCR for the ITS1 spacer
The S. lupi ITS1 spacer was characterized from four adult S. lupi specimens using primers rDNA2 [13] and rDNA1.58S [14], following a previously described protocol [14]. Then, the amplified sequence was inserted into pCR2-TOPO vectors (Thermo Fisher Scientific Inc.) and cloned. Specific primers were designed using Primer-BLAST [15], to amplify a 135 bp fragment within the ITS1 of S. lupi. The HRM qPCR assay used primers SlITS-F (5′-AAA CGG TGT CCC ATG TTG G-3′) and SlITS-R (5′-GCA GCA CAA TAG CTT GAC GC-3′) at a final concentration of 500 nM. Each reaction consisted of 1 μl of each primer, 0.6 μl of SYTO-9 (Invitrogen, California, USA), 4.4 μl of PCR grade water (Biological Industries Inc.), 10 μl of Maxima HotStart Master Mix® (Thermo Fisher Scientific Inc.) and 3 μl of DNA. The amplification reaction consisted of an initial hold at 95 °C for 4 min, followed by 45 cycles of 95 °C for 5 s, 60 °C for 15 s and 72 °C for 10 s. A melt curve from 80 °C to 95 °C was constructed with increments of 1 °C/s, followed by a hybridization step from 90 °C to 50 °C. An HRM curve was constructed from 70 °C to 85 °C, with 0.1 °C/s increments.
HRM qPCR for the 18S locus
Primers for a fragment of the 18S locus of S. lupi available in GenBank (HQ674750.1) were designed using Primer-BLAST [15]. Accordingly, primers Sl18S-F (5′-AAG CTC CGA CTT TTG GAC GA-3′) and Sl18S-R (5′-GTC ACT ACC TCC TCA TGC CG-3′) were constructed to amplify a 270 bp fragment of the 18S gene and used at a final concentration of 500 nM. The reactions consisted of 1 μl of each primer, 0.6 μl of SYTO-9 (Invitrogen), 4.4 μl of PCR grade water (Biological Industries Inc.), 10 μl of Maxima HotStart Master Mix® (Thermo Fisher Scientific Inc.) and 3 μl of DNA. The amplification reaction consisted of an initial hold of 95 °C for 4 min, followed by 50 cycles of 95 °C for 5 s, 59 °C for 15 s and 72 °C for 10 s. A melt curve from 80 °C to 95 °C was constructed with 1 °C/s increments, followed by a hybridization step from 90 °C to 50 °C. An HRM was constructed from 80 °C to 90 °C with 0.1 °C/s increments.
HRM qPCR for the cytb locus
A sequence of the cytb gene from GenBank (KC305876.1) [16] was used for the development of novel primers for this locus. Hence, primers Slcytb-F (5′-ACT GCG GGG GAG TCT TTC T-3′) and Slcytb-R (5′-AGT AAT AAC AAC CGC CGC CC-3′) were designed to amplify a 260 bp fragment of the cytb gene. Primers were used at a final concentration of 250 nM. Each reaction consisted of 0.5 μl of each primer, 0.6 μl of SYTO-9 (Invitrogen), 5.4 μl of PCR grade water (Biological Industries Inc.), 10 μl of Maxima HotStart Master Mix® (Thermo Fisher Scientific Inc.) and 3 μl of DNA. The program consisted of an initial hold at 95 °C for 4 min and 45 cycles of 95 °C for 5 s, 57 °C for 15 s and 72 °C for 5 s. A melt curve from 75 °C to 90 °C was constructed, with 1 °C/s increments, followed by a hybridization step. The HRM curve was constructed from 75 °C to 90 °C with 0.1 °C/s increments.
All HRM qPCRs were run in the Rotor Gene 6000 Cycler (Qiagen, Hilden, Germany). Each reaction was run with positive, negative and non-template controls (NTC). Reaction controls included S. lupi adult-DNA (positive control), DNA from a S. lupi-negative dog fecal sample (negative control, determined by fecal flotation in three consecutive samples), a point of the standard curve by triplicate and NTC with PCR-grade water.
Semi-nested PCR for detection of the S. lupi cox1 gene
The semi-nested PCR for the amplification of a fragment of the cox1 gene of S. lupi was performed as described previously [5], with the following modifications to improve the detection of S. lupi DNA, based on preliminary tests. In the first round of amplification, primers NTF and NTR were employed at a final concentration of 200 nM instead of 2 pmol, while MgCl2 was used at a final concentration of 3 mM, and finally, 3 μl of DNA standard, were added to each reaction in PCR-ready Syntezza® tubes (Syntezza Bioscience Ltd., Jerusalem, Israel). In the nested PCR reaction, the SlInt and NTR primers were used at a final concentration of 200 nM, instead of 2 pmol, while MgCl2 was used at a final concentration of 3 mM. DNA from the first PCR reaction was diluted 40-fold in PCR-grade water, and 5 μl of this dilution was employed as a template. The PCR cycling protocol consisted of an initial hold at 95 °C for 5 min, followed by 40 cycles of 94 °C for 45 s, 54 °C or 60 °C for 1 min in the primary and secondary reaction, respectively, and 72 °C for 1 min, and a final elongation at 72 °C for 5 min. The fecal samples from the dogs with spirocercosis were run together with S. lupi adult-DNA, a S. lupi-negative dog fecal control and an NTC with PCR-grade water. Additionally, DNA standards of S. lupi were run in triplicate to estimate the assay detection limit.
Experimental controls
DNA from the following gastrointestinal parasites was used as controls to evaluate the possible cross-amplification of the S. lupi HRM qPCR assays: Ancylostoma caninum, Calodium hepaticum, Strongyloides stercoralis, Toxocara canis, Toxascaris leonina, Taenia solium, Dyphillobothrium latum, Dipylidium caninum, Spirometra mansoni, Echinococcus granulosus, Echinococcus multilocularis, Giardia duodenalis, Neospora caninum and Cystoisospora canis. The life stages, sources and DNA-extraction methods of each of these parasites are indicated in Additional file 1: Table S1. Additionally, an experimental control mimicking severe toxocariasis infection with T. canis [17] was prepared. Briefly, a concentrated egg suspension of T. canis was obtained from the uterus of four adult female T. canis worms. The eggs were counted in triplicate, and their concentration adjusted to 93,000 epg.
Sequence analysis
Positive DNA amplicons were purified (Exo-SAP New England Bio-Labs Inc., Ipswich, USA) and sequenced using the Big-Dye Terminator cycle sequencing chemistry from Applied Biosystems ABI3700 DNA Analyzer and the ABI’s Data Collection and Sequence Analysis software (ABI Carlsbad, USA). Samples were considered positive for S. lupi when the obtained amplicon had at least 97% sequence identity, and the closest match was to S. lupi sequences available in GenBank using the BLAST 2.2.28 program.
Data analysis
Each positive sample was run in triplicate, to calculate the geometric mean of S. lupi epg and standard deviation. Fecal samples were considered positive when all three replicates were amplified. The HRM qPCRs Cq values of each positive sample were used to estimate the S. lupi epg, as in the following formula:
Cq stands for quantification cycle, b for y-intercept, m for the slope obtained in the standard curve, and Cf is a correction factor, calculated for each fecal sample. The average Cq value of each positive sample was interpolated in the standard curve equation to obtain the epg (first factor of the equation). The variability in the mass of fecal sample employed for DNA extraction was normalized by dividing 0.2 by the exact weight of the extracted fecal sample (in g).
Statistical analysis
Cohen’s kappa coefficient was calculated to determine the agreement in the detection of spirocercosis cases between the four molecular assays and the fecal flotation. Κ-values were interpreted according to Landis & Koch [18]. The Wilcoxon signed rank test was used to determine the differences in the epg concentration in the fecal samples of the dogs with spirocercosis obtained by fecal flotation and the estimated values from the HRM qPCRs. This same test was used for comparing the epg obtained in the HRM qPCRs for the ITS1 and 18S. Spearman’s ranks correlation coefficient was calculated to determine the correlation between the epg as quantified by the ITS1 and 18S HRM qPCRs. Lastly, Bonferroni’s correction was applied for multiple comparisons. Significance was considered when P < 0.017. Statistical analyses were performed using the SPSS 22.0 software package (IBM Corp., USA).
Results
HRM real time qPCR assays performance
The standard curve for the ITS1, 18S and cytb HRM qPCR assays showed efficiencies of 90%, 103% and 134%, respectively. The y-intercept, slope and linear correlation (R2) are shown in Table 1.
The HRM qPCRs for the ITS1 and 18S loci showed detection limits of 0.2 epg. The HRM qPCR for the cytb detected down to 6 epg. In contrast, the detection limit of the cox1 semi-nested PCR with the employed modifications was 526 epg (Additional file 2: Figure S1). The designed primers for ITS1 did not cross-amplify any DNA from the other examined common gastrointestinal dog parasites. The HRM qPCR for the 18S gene amplified DNA from T. canis and T. leonina after 30 cycles. Dipylidium caninum DNA was amplified in the HRM qPCR for cytb but only after cycle 30.
Detection of spirocercosis
The molecular methods targeting ITS1 and 18S of S. lupi detected higher numbers of positive fecal samples than the currently available diagnostic methods (i.e. fecal flotation and semi-nested cox1 PCR). The HRM qPCR for ITS1 and 18S were positive in 61.1% (11/18; 95% CI: 35.7–82.7%) and 44.4% (8/18; 95% CI: 21.5–69.2%) of the fecal samples from the dogs with spirocercosis, respectively. The fecal flotation assay identified S. lupi eggs in 27.8% of the dogs (5/18; 95% CI: 9.7–53.5%), while the HRM qPCR for cytb and the semi-nested cox1 PCR identified only two and one samples as positive, respectively.
Agreement for the detection of positive and negative samples for S. lupi was observed between the methods. There were fair (κ = 0.39) and good (κ = 0.65) agreements between the HRM qPCR for ITS1 and the fecal flotation, and between the HRM qPCR for 18S and the fecal flotation, respectively. The HRM qPCRs for ITS1 and 18S were positive in all the fecal samples found to have S. lupi eggs by the fecal flotation (Table 2). Furthermore, 6 and 3 samples from dogs with spirocercosis detected positive by the HRM qPCR for ITS1 and 18S, respectively, were negative by the fecal flotation. The fecal flotation results had a moderate agreement with the cytb HRM qPCR (κ = 0.50) and fair agreement with the semi-nested PCR for the cox1 (κ = 0.26). The semi-nested cox1 PCR and the HRM cytb qPCR assay were positive in 10% and 20%, respectively, of the samples detected as positive by the fecal flotation. When comparing the HRM qPCRs for ITS1 and 18S, their agreement was good (κ = 0.67). There was none to slight agreement when comparing the results of ITS1 HRM qPCR and cytb HRM qPCR (κ = 0.15) and of the semi-nested cox1 PCR (κ = 0.16), as well as between the 18S HRM qPCR and the semi-nested cox1 PCR (κ = 0.14).
Repeated oesophagoscopies of the dogs included in this study indicated that the oesophageal nodules in all 18 dogs had resolved after 16-20 weeks of treatment, proving that spirocercosis was benign.
Quantification of S. lupi eggs from fecal samples
Quantification of S. lupi eggs was successfully achieved by the HRM qPCRs for ITS1, 18S and cytb loci, and by the fecal flotation (Table 2). No significant difference in the calculated concentrations was found between the results when all these assays were compared (Wilcoxon signed rank test; P > 0.05 for all). However, when comparing the eggs quantified by the HRM qPCR for ITS1 and 18S, there was a significant difference (Wilcoxon signed rank test, W = -2.803, P = 0.005) with a significant positive correlation (Spearman’s r s = 0.918; P < 0.0001; Additional file 3: Figure S2).
Sequence analysis
All fecal samples found positive by the HRM qPCRs for 18S, and cytb loci were identified as containing S. lupi DNA by sequence comparison to publicly available sequences on the GenBank database. The accession number with the closest match and percentage of identity in the 18S HRM qPCR was HQ674750.1 (99%), and for the cytb qPCR and cox1 semi-nested PCR, it was KC305876.1 (97% and 96%, respectively). No S. lupi ITS1 sequence has been deposited in GenBank, and the closest match was Cylicospirura petrowi (KM434335.1, 96% identity with 47% coverage). Representative S. lupi ITS1, 18S, cytB and cox1 sequences obtained in this study were deposited in GenBank under accession numbers MF425539, MF79497, MF804403 and MF804404, respectively.
Discussion
The current diagnosis of spirocercosis is challenging in clinical practice due to the variable nature of the clinical signs related to the different clinical presentations, parasite migration and development within the host, and the intermittent, unpredictable shedding of eggs in the feces [1]. Thus, to assess and improve the diagnosis of oesophageal spirocercosis, the present study explored the use of HRM qPCR assays to detect and quantify fecal S. lupi eggs. Accordingly, three novel HRM qPCRs were designed, and fecal samples from dogs confirmed with typical S. lupi oesophageal nodules were tested and compared with two currently available methods, namely, conventional coproscopy by the fecal flotation [7] and the cox1 semi-nested PCR [5]. The detection of fecal S. lupi DNA of dogs with oesophageal spirocercosis was successful and efficiently achieved in the HRM qPCRs targeting ITS1 and 18S.
The ribosomal (rDNA) and mitochondrial (mtDNA) DNA are among the most frequently used targets for PCR, due to their high copy number and their ability to display nucleotide sequence variability among species, which facilitates identification of parasite species [19]. The three molecular protocols described in this study, two associated with rDNA and one with mtDNA, successfully amplified S. lupi DNA. The HRM qPCR designed for ITS1 demonstrated the best potential for the diagnosis of S. lupi DNA and did not cross-amplify DNA of other gastrointestinal parasites, as compared to the 18S and cytb assays. The ITS1 and 18S assays exhibited the same detection limit (0.2 epg), in contrast to the cytb assay which showed a 30-fold higher minimal detection limit (6 epg). The diagnostic utility of the ITS1 and 18S assays might be explained by a high loci copy number or good sequence homology to the primers used and better reaction conditions. Nevertheless, the ITS1 assay detected the most spirocercosis cases in this study (Table 2). In the case of the cytb assay, several primer pairs and conditions were tested, but the sensitivity of this assay could not be further improved.
The currently available methods, fecal flotation and the semi-nested cox1 PCR, demonstrated a much lower performance than the molecular methods described in this study, with the exception of the cytb assay. The higher performance of the ITS1 and 18S assays in comparison to the fecal flotation likely results from the superior sensitivity of DNA-detection methods compared to microscopic methods, which require the detection of intact eggs. A similar scenario was observed in the detection of microfilariae DNA by PCR when compared with whole microfilariae microscopic identification in canine blood samples [20]. The HRM qPCRs developed for this study may be able to detect DNA from disrupted eggs, worm tissue parts or worm cellular DNA discharged into the host’s gastrointestinal lumen. Lastly, the molecular methods use DNA extracted from a fecal volume which is 15-fold smaller than that required for the fecal flotation. Nevertheless, the ITS1 and 18S HRM qPCR assays identified all the cases found positive by fecal flotation, as well as additional ones that were negative by fecal flotation but were positive by oesophagoscopy.
The semi-nested PCR had the highest detection limit in this study. To date, this was the only molecular assay described for detecting fecal S. lupi DNA [5]. However, nested-PCR protocols are usually more time consuming, requiring two reactions, and are prone to contamination, as reported in the diagnosis of viruses and bacteria of clinical importance [9, 10]. Therefore, the protocols designed herein demonstrated superior laboratory efficiency, requiring only a single reaction and being more effective in detecting spirocercosis than the nested PCR.
Oesophagoscopy is considered the most sensitive and specific diagnostic technique for the detection of oesophageal spirocercosis [21], but is limited by high cost, availability of specialized equipment, requirement for general anesthesia, potential complications (e.g. oesophageal bleeding or rupture) and the need for clinical expertise for the accurate identification of S. lupi nodules [1, 5]. Although in the present study, up to 61% of the spirocercosis cases were detected as positive by the ITS1 HRM qPCR, seven cases verified by oesophagoscopy were not detected as positive by any other methodology. This might be explained by the intermittent egg shedding into the feces, and low fecal egg numbers, which might have been absent in the small volume of fecal sample obtained for DNA extraction, or single-sex worm oesophageal infection, resulting in the absence of egg shedding [22]. Thus, we recommend analyzing a second fecal sample, obtained a few days after a negative result, since this can increase the positivity of detection in fecal samples, as previously suggested [21, 22].
The new HRM qPCRs might be helpful not only for detecting and quantifying fecal S. lupi but also for monitoring therapy and its effectiveness, as evident by the decrease and eventual absence of fecal S. lupi eggs and DNA in a previous study [23]. Previous studies in experimentally infected dogs have shown that fecal samples became negative for S. lupi eggs within 3 to 10 days after the first or second doramectin dose, given at a 2-week interval [23]. In naturally infected dogs, it has been shown that fecal samples remain positive for S. lupi eggs for up to 4 weeks after treatment with levamisole hydrochloride [24]. Therefore, the new molecular techniques, which have demonstrated the ability to detect very small amounts of fecal S. lupi DNA, equivalent to less than one epg, should be considerably more sensitive for detecting persistent infection than conventional coproscopy.
Interestingly, the HRM qPCR for ITS1 quantified a higher number of eggs than the assay for 18S (Table 2 and Additional file 3: Figure S2), although these loci are located in the same ribosomal operon, and should, therefore, be present in identical numbers of copies. This discrepancy might result from a higher annealing specificity of the primers to their target sequence in the case of the ITS1.
The HRM qPCR for ITS1 has amplified S. lupi DNA exclusively, whereas the 18S and cytb HRM qPCRs also amplified DNA from other canine gastrointestinal parasites. The cross-amplification of T. canis and T. leonina by the 18S HRM qPCR might be explained by a sequence similarity between the two nematodes, and because for T. canis, DNA from a very high egg concentration was employed, to simulate severe infection [25]. Since the ITS1 HRM qPCR was the only assay which did not cross-amplify other parasite DNA, this assay would be the PCR method of choice for the diagnosis of S. lupi in fecal samples.
Conclusions
This study describes novel and efficient molecular assays for the detection of S. lupi DNA in feces from dogs with oesophageal spirocercosis and the quantification of fecal egg loads. The ITS-1 HRM qPCR showed the best performance and was superior to fecal flotation and other PCR assays. These new molecular assays are potential tools for the evaluation of treatment success and prognosis of spirocercosis in conjunction with clinical evaluation and the employment of imaging techniques.
Abbreviations
- 18S:
-
ribosomal subunit 18S
- Cq:
-
quantification cycle
- cytb :
-
cytochrome oxidase b
- EPG:
-
Eggs per gram
- HRM:
-
High-resolution melt
- ITS1:
-
Internal transcribed spacer 1
- NTC:
-
Non-template control
- PCR:
-
Polymerase chain reaction
- qPCR:
-
quantitative polymerase chain reaction
References
van der Merwe LL, Kirberger RM, Clift S, Williams M, Keller N, Naidoo V. Spirocerca lupi infection in the dog: a review. Vet J. 2008;176(3):294–309.
Mukaratirwa S, Singh VP. Prevalence of gastrointestinal parasites of stray dogs impounded by the Society for the Prevention of Cruelty to Animals (SPCA), Durban and Coast, South Africa. J S Afr Vet Assoc. 2010;81:123–5.
Gholi-Toluei M, Amniattalab A, Rasouli S. Oesophageal spirocercosis in stray dogs of Urmia. Res Op An Vet Sci. 2015;5:388–91.
Aroch I, Markovics A, Mazaki-Tovi M, Kuzi S, Harrus S, Yas E, et al. Spirocercosis in dogs in Israel: a retrospective case-control study (2004–2009). Vet Parasitol. 2015;211:234–40.
Traversa D, Avolio S, Modrý D, Otranto D, Iorio R, Aroch I, et al. Copromicroscopic and molecular assays for the detection of cancer-causing parasitic nematode Spirocerca lupi. Vet Parasitol. 2008;157:108–16.
Aroch I, Rojas A, Slon P, Lavy E, Segev G, Baneth G. Serological cross-reactivity of three commercial in-house immunoassays for detection of Dirofilaria immitis antigens with Spirocerca lupi in dogs with benign oesophageal spirocercosis. Vet Parasitol. 2015;211:303–5.
Markovics A, Medinski B. Improved diagnosis of low intensity Spirocerca lupi infection by the sugar flotation method. J Vet Diagn Investig. 1996;8:400–1.
Coskun SZ. Diagnosis of Spirocerca lupi by IFAT in naturally infected dogs. Türk Parazitol Derg. 1995;19:541–9.
Lam WY, Yeung AC, Tang JW, Ip M, Chan EW, Hui M, et al. Rapid multiplex nested PCR for detection of respiratory viruses. J Clin Microbiol. 2007;45:3631–40.
Yamamoto Y. PCR in diagnosis of infection: detection of bacteria in cerebrospinal fluids. Clin Diagn Lab Immunol. 2002;9:508–14.
Gasser RB. Molecular tools - advances, opportunities and prospects. Vet Parasitol. 2006;136:69–89.
Rojas A, Freedberg N, Markovics A, Gottlieb Y, Baneth G. Influence of physical and chemical factors on the embryonation, hatching and infectivity of Spirocerca lupi. Vet Parasitol. 2017;242:71–8.
Vrain T, Wakarchuk D, Lévesque A, Hamilton R. Intraspecific rDNA restriction fragment length polymorphism in the Xiphinema americanum group. Fund Appl Nematol. 1992;15:563–73.
Cherry T, Szalanski AL, Todd TC, Powers TO. The internal transcribed spacer region of Belonolaimus (Nemata: Belonolaimidae). J Nematol. 1997;29:23–9.
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134.
Liu GH, Wang Y, Song HQ, Li MW, Ai L, Yu XL, et al. Characterization of the complete mitochondrial genome of Spirocerca lupi: sequence, gene organization and phylogenetic implications. Parasit Vectors. 2013;6:45.
Jacobs DE, Fisher MA. Recent developments in the chemotherapy of Toxocara canisinfection in puppies and the prevention of toxocariasis. In: Lewis JW, Maizels RM, editors. Toxocara and toxocariasis: clinical, epidemiological and molecular perspectives. London: Institute of Biology; 1993. p. 111–6.
Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–74.
Verweij JJ, Stensvold CR. Molecular testing for clinical diagnosis and epidemiological investigations of intestinal parasitic infections. Clin Microbiol Rev. 2014;27:371–418.
Rojas A, Rojas D, Montenegro VM, Baneth G. Detection of Dirofilaria immitis and other arthropod-borne filarioids by an HRM real-time qPCR, blood-concentrating techniques and a serological assay in dogs from Costa Rica. Parasit Vectors. 2015;8:170.
Mazaki-Tovi M, Baneth G, Aroch I, Harrus S, Kass PH, Ben-Ari T, et al. Canine spirocercosis: clinical, diagnostic, pathologic, and epidemiologic characteristics. Vet Parasitol. 2002;107:235–50.
Christie J, Schwan EV, Bodenstein LL, Sommerville JE, van der Merwe LL. The sensitivity of direct faecal examination, direct faecal flotation, modified centrifugal faecal flotation and centrifugal sedimentation/flotation in the diagnosis of canine spirocercosis. J S Afr Vet Assoc. 2011;82:71–5.
Lavy E, Aroch I, Bark H, Markovics A, Aizenberg I, Mazaki-Tovi M, et al. Evaluation of doramectin for the treatment of experimental canine spirocercosis. Vet Parasitol. 2002;109:65–73.
Giannelli A, Baldassarre V, Ramos RA, Lia RP, Furlanello T, Trotta M, et al. Spirocerca lupi infection in a dog from southern Italy: an "old fashioned" disease? Parasitol Res. 2014;113:2391–4.
Durant JF, Irenge LM, Fogt-Wyrwas R, Dumont C, Doucet JP, Mignon B, et al. Duplex quantitative real-time PCR assay for the detection and discrimination of the eggs of Toxocara canis and Toxocara cati (Nematoda, Ascaridoidea) in soil and fecal samples. Parasit Vectors. 2012;5:288.
Acknowledgements
The authors thank Dr. Victor Montenegro, Dr. Adrian Montero, Dr. Paul Slon and Dr. Harold Salant for their kind provision of parasite controls.
Funding
The study was supported by internal funding of Prof. Gad Baneth’s laboratory, and a stipend from the University of Costa Rica granted to Alicia Rojas.
Availability of data and materials
Representative S. lupi ITS1, 18S, cytb and cox1 sequences obtained in this study were deposited in GenBank under accession numbers MF425539, MF79497, MF804403 and MF804404, respectively.
Author information
Authors and Affiliations
Contributions
AR and GB conceived the study, analyzed the data and drafted the manuscript. AM, GS and IA, diagnosed and treated the dogs with spirocercosis, collected the fecal samples from dogs and S. lupi adult specimens, and took part in drafting the manuscript. AM performed the flotation method of all fecal samples. AR grew the adult worms in vitro, performed DNA extraction and PCR. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study was conducted according to the Hebrew University experiments in animal policy and approved by the Hebrew University’s Koret School of Veterinary Medicine Veterinary Teaching Hospital Internal Ethics Review Committee approval number KSVM-VTH/21_2014.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1: Table S1.
DNA controls of dog parasites used in the study to test the specificity of the HRM qPCRs targeting different loci of S. lupi. (DOCX 12 kb)
Additional file 2: Figure S1.
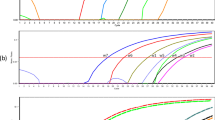
Limit of detection of the cox1 semi-nested PCR for S. lupi. DNA-standards with known epg concentration were run by triplicates, and the detection limit was estimated as the last concentration of which more than 67% of the samples were still positive. (PNG 867 kb)
Additional file 3: Figure S2.
Correlation in the eggs per gram quantified in the HRM qPCRs for the ITS1 and 18S of S. lupi in fecal samples from dogs with spirocercosis. Logarithmic scales are used in both x- and y-axes. (PNG 16 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Rojas, A., Segev, G., Markovics, A. et al. Detection and quantification of Spirocerca lupi by HRM qPCR in fecal samples from dogs with spirocercosis. Parasites Vectors 10, 435 (2017). https://doi.org/10.1186/s13071-017-2374-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-017-2374-3