Abstract
Background
Clostridium thermocellum is a cellulolytic anaerobic thermophile that is a promising candidate for consolidated bioprocessing of lignocellulosic biomass into biofuels such as ethanol. It was previously shown that expressing Thermoanaerobacterium saccharolyticum adhA in C. thermocellum increases ethanol yield.In this study, we investigated expression of adhA genes from different organisms in Clostridium thermocellum.
Methods
Based on sequence identity to T. saccharolyticum adhA, we chose adhA genes from 10 other organisms: Clostridium botulinum, Methanocaldococcus bathoardescens, Thermoanaerobacterium ethanolicus, Thermoanaerobacter mathranii, Thermococcus strain AN1, Thermoanaerobacterium thermosaccharolyticum, Caldicellulosiruptor saccharolyticus, Fervidobacterium nodosum, Marinitoga piezophila, and Thermotoga petrophila. All 11 adhA genes (including T. saccharolyticum adhA) were expressed in C. thermocellum and fermentation end products were analyzed.
Results
All 11 adhA genes increased C. thermocellum ethanol yield compared to the empty-vector control. C. botulinum and T. ethanolicus adhA genes generated significantly higher ethanol yield than T. saccharolyticum adhA.
Conclusion
Our results indicated that expressing adhA is an effective method of increasing ethanol yield in wild-type C. thermocellum, and that this appears to be a general property of adhA genes.
Similar content being viewed by others
Background
Clostridium thermocellum is a cellulolytic anaerobic thermophile that is considered to be a promising candidate for consolidated bioprocessing of lignocellulosic biomass, into biofuels such as ethanol, due to its native ability to solubilize lignocellulose [1]. A key limitation of this organism is that it produces ethanol only at low yield (20% of the theoretical maximum) [2]. Strategies to increase ethanol yield in C. thermocellum include deleting the pathways for acetic acid, lactic acid, and hydrogen production [3,4,5,6], and introducing heterologous genes from ethanol production pathways in other organisms [2, 7], such as Thermoanaerobacterium saccharolyticum. Recently, it was shown that AdhA plays an important role in ethanol production in strains of T. saccharolyticum engineered for homoethanol production [8]. This enzyme was subsequently expressed in C. thermocellum and shown to increase ethanol yield and titer by 40% [3]. In this study, we chose adhA genes from 10 additional organisms, expressed them in C. thermocellum and observed the effect on ethanol production.
Methods
Plasmid and strain construction
Plasmids used for adhA expression in C. thermocellum are listed in Table 1. Plasmids were constructed based on the C. thermocellum expression plasmid pDGO144 as previously described [9]. The Clo1313_2638 promoter [9] and adhA gene were cloned into the HindIII site of pDGO144 using standard molecular biology techniques. The correct reading frame and sequence of each adhA gene in the resulting plasmids in Table 1 were confirmed by Sanger Sequencing (GENEWIZ). Complex medium CTFÜD [10] was used to culture wild-type C. thermocellum. Plasmids expressing adhA genes were transformed into wild-type C. thermocellum using the transformation protocol as previously described [10]. Selection was carried out using thiamphenicol at a final concentration of 6 µg/ml. Single colonies were picked and re-inoculated into CTFÜD medium containing 6 µg/ml thiamphenicol; cultures were saved for further analysis. The presence of adhA genes in the cultures was confirmed by PCR. Primers used for the confirmation are Fwd: GACGAAAAAGCCGATGAAG, Rev: CCTTTTTTAAAAGTCAATCCCG. The size of the PCR product was used to confirm adhA insertion: the PCR product of the empty vector is 178 bp, and the PCR product containing the adhA gene insertion is ~ 1400 bp (with slight variation due to differences in lengths of the adhA genes).
Fermentations and end-product analysis
For fermentation end-product analysis, strains were transferred three times in defined MTC-5 medium [11] with 4.7 g/l cellobiose at 1% inoculum (v/v). End-product measurements were taken on the 3rd transfer. Cultures were grown in Corning™ Falcon™ 15 ml Conical Centrifuge Tubes and incubated anaerobically without shaking at 55 °C for 72 h. Upon harvesting, cultures were prepared as previously described for HPLC (High-Pressure Liquid Chromatography) analysis [8]. Ethanol yield was calculated as the percentage of theoretical yield based on the amount of ethanol produced and substrate consumed: \(\left[ {{\text{Yield ethanol }}\left( \%{{\text{ maximum theroretical}}} \right) = \frac{{\rm Amount of ethanol produced (mM)}}{4*{\rm Amount of cellobiose consumed (mM)}}} \right]\). Carbon balance was calculated based on the fermentation products measured as previously described [12]: \(\left[ {{\text{Carbon balance }}\left( {\text{\% }} \right) = \frac{{\left[ {\rm Acetate} \right] + \left[ {\rm Ethanol} \right] + \left[ {\rm Lactate} \right]\left( {\rm mM} \right)}}{{4*\left[ {\rm cellobiose consumed} \right]\left( {\rm mM} \right)}}} \right].\)
Phylogenetic analysis
The amino acid sequences of different AdhA proteins were aligned using CLC Main Workbench 7.7.3, and a phylogenetic tree was created using the Neighbor Joining algorithm. Distance is expressed as substitutions per 100 amino acids; multiple substitutions at the same site were corrected for using the Kimura method. Bootstrap analysis was performed with 1000 replicates.
Results and discussion
adhA genes from different organisms
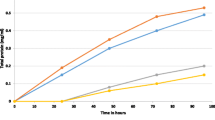
Sequences with homology to the T. saccharolyticum AdhA were searched using the BLAST (Basic Local Alignment Search Tool) algorithm [13]. AdhA sequences from different organisms were chosen based on protein sequence identity to T. saccharolyticum AdhA, with an identity range of 57–90% (Table 1). Most of the selected organisms were thermophilic bacteria with an optimal growth temperature greater than 50 °C as presented in Table 1. Clostridium botulinum, a mesophilic bacterium that grows at 37 °C, was also chosen with the intention of exploring the heat stability of AdhA. A phylogenetic tree of AdhA proteins used in this study is presented in Fig. 1.
Phylogenetic tree of the AdhA proteins and ethanol yield for 11 C. thermocellum strains expressing adhA genes from different organisms. a The distance-based phylogenetic tree generated from the alignments of the AdhA proteins. b Ethanol yield for all of the adhA-expressing strains. Strains were cultured in MTC-5 medium containing 4.7 g/l cellobiose and 6 μg/ml thiamphenicol at 55 °C for 72 h. The maximum theoretical yield is 4 mol of ethanol per mole of cellobiose consumed. Data were collected from triplicate experiments. Error bars represent one standard deviation. Wild-type C. thermocellum expressing the empty vector pDGO144 (LL1535) was used as negative control (indicated in red), and the strain expressing T. saccharolyticum adhA was used as positive control (indicated in green). All of the other 10 strains were shown as the experimental group (indicated in gray). Ethanol yields of experimental group strains were compared to the positive control using a two-tailed unpaired t test, and p values are reported where significant
Fermentation behavior of C. thermocellum strains expressing different adhA genes
The 11 adhA genes described above, including T. saccharolyticum adhA, were cloned into expression plasmid pDGO144 and expressed in wild-type C. thermocellum. Fermentation results for all of the strains are presented in Table 2. Wild-type C. thermocellum harboring the empty pDGO144 plasmid was used as a negative control strain. Ethanol yield was calculated based on the amount of ethanol produced from the amount of cellobiose consumed. Two-tailed unpaired T tests were performed on the ethanol yields of the strains with three biological replicates to assess statistical significance. To evaluate the effect of expressing adhA genes in C. thermocellum, ethanol yield for each strain was compared to the empty vector negative control. The strain expressing T. saccharolyticum adhA, LL1536, had significantly higher ethanol yield than the empty vector control (p < 0.0001), agreeing with previous results [2]. The other 10 strains expressing adhA genes all had significantly higher ethanol yield compared to the empty-vector control strain (p < 0.05). When compared to the positive control that expressed T. saccharolyticum adhA (LL1536), two strains exhibited significantly higher ethanol yield: Strain LL1527 expressing C. botulinum adhA (p = 0.0001) and strain LL1526 expressing T. ethanolicus adhA (p = 0.0353) (Fig. 1). The top two AdhAs in terms of increasing ethanol yield appeared to be evolutionarily distant from each other: C. botulinum and T. ethanolicus, and we did not observe any correlation between sequence similarity and effect on ethanol production. In general, most of the additional ethanol production came at the expense of acetate production (Table 2). This is consistent with other reports indicating that there appears to be an oversupply of NADPH in C. thermocellum [14, 15], and that this can be used to divert C2 flux (i.e., acetyl-CoA) to ethanol in the presence of an NADPH-linked ADH enzyme [8, 9, 16]. Lactate and malate were minor fermentation products. Carbon balances were calculated based on the fermentation end products measured in this study, and they were generally 65–75% closed. The remaining 25–35% of the substrate carbon is likely present in biomass or un-measured fermentation products such as amino acids.
Conclusions
Our results indicate that expressing adhA is an effective method of increasing ethanol yield in wild-type C. thermocellum, and that this appears to be a general property of adhA genes, rather than a property specific to the adhA gene from T. saccharolyticum. Although most of the adhAs studied in this work are from thermophiles, the largest increase in ethanol production came from the adhA gene from C. botulinum, a mesophile with an optimal growth temperature of 37 °C.
References
Olson DG, McBride JE, Joe Shaw A, Lynd LR. Recent progress in consolidated bioprocessing. Curr Opin Biotechnol. 2012;23:396–405. doi:10.1016/j.copbio.2011.11.026.
Hon S, Olson DG, Holwerda EK, Lanahan AA, Murphy SJL, Maloney MI, et al. The ethanol pathway from Thermoanaerobacterium saccharolyticum improves ethanol production in Clostridium thermocellum. Metab Eng. 2017;42:175–84. doi:10.1016/j.ymben.2017.06.011.
Argyros DA, Tripathi SA, Barrett TF, Rogers SR, Feinberg LF, Olson DG, et al. High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes. Appl Environ Microbiol. 2011;77:8288–94.
Rydzak T, Lynd LR, Guss AM. Elimination of formate production in Clostridium thermocellum. J Ind Microbiol Biotechnol. 2015;42:1263–72.
Biswas R, Wilson CM, Zheng T, Giannone RJ, Dawn M. Elimination of hydrogenase active site assembly blocks H2 production and increases ethanol yield in Clostridium thermocellum. Biotechnol Biofuels. 2015;8:20.
Papanek B, Biswas R, Rydzak T, Guss AM. Elimination of metabolic pathways to all traditional fermentation products increases ethanol yields in Clostridium thermocellum. Metab Eng. 2015;32:49–54. doi:10.1016/j.ymben.2015.09.002.
Background EA, Benziman M, Russo A, Hochman S, Weinhouse H, Bisswanger H, et al. Redirecting carbon flux through exogenous pyruvate kinase to achieve high ethanol yields in Clostridium thermocellum. Metab Eng. 2013;15:151–8. doi:10.1016/j.ymben.2012.11.006.
Zheng T, Olson DG, Murphy SJ-L, Shao X, Tian L, Lynd LR. Both adhE and a separate NADPH-dependent alcohol dehydrogenase gene, adhA, are necessary for high ethanol production in Thermoanaerobacterium saccharolyticum. J Bacteriol. 2017;199:1–10.
Hon S, Lanahan AA, Tian L, Giannone RJ, Hettich RL, Olson DG, et al. Development of a plasmid-based expression system in Clostridium thermocellum and its use to screen heterologous expression of bifunctional alcohol dehydrogenases (adhEs). Metab Eng Commun. 2016;3:120–9.
Olson DG, Lynd LR. Transformation of Clostridium thermocellum by electroporation. Methods Enzymol. 2012;510:317–30.
Zhang YH, Lynd LR. Quantification of cell and cellulase mass concentrations during anaerobic cellulose fermentation: development of an enzyme-linked immunosorb, ient assay-based method with application to Clostridium thermocellum batch cultures. Anal Chem. 2003;75:219–27.
Ellis LD, Holwerda EK, Hogsett D, Rogers S, Shao X, Tschaplinski T, et al. Closing the carbon balance for fermentation by Clostridium thermocellum (ATCC 27405). Bioresour Technol. 2012;103:293–9. doi:10.1016/j.biortech.2011.09.128.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
van der Veen D, Lo J, Brown SD, Johnson CM, Tschaplinski TJ, Martin M, et al. Characterization of Clostridium thermocellum strains with disrupted fermentation end-product pathways. J Ind Microbiol Biotechnol. 2013;40:725–34.
Olson DG, Hörl M, Fuhrer T, Cui J, Zhou J, Maloney MI, et al. Glycolysis without pyruvate kinase in Clostridium thermocellum. Metab Eng. 2016;2016(39):169–80.
Zheng T, Olson DG, Tian L, Bomble YJ, Himmel ME, Lo J, et al. Cofactor specificity of the bifunctional alcohol and aldehyde dehydrogenase (AdhE) in wild-type and mutants of Clostridium thermocellum and Thermoanaerobacterium saccharolyticum. J Bacteriol. 2015;197:2610–9. doi:10.1128/JB.00232-15.
Klinke H, Thomsen A, Ahring B. Potential inhibitors from wet oxidation of wheat straw and their effect on growth and ethanol production by Thermoanaerobacter mathranii. Appl Microbiol Biotechnol. 2001;57:631–8.
Pei J, Zhou Q, Jiang Y, Le Y, Li H, Shao W, et al. Thermoanaerobacter spp. control ethanol pathway via transcriptional regulation and versatility of key enzymes. Metab Eng. 2010;12:420–8. doi:10.1016/j.ymben.2010.06.001.
Tanner FW, Oglesby EW. Influence of temperature on growth and toxin production by Clostridium botulinum. Food Res. 1936;1:481–94.
Li D, Stevenson KJ. Alcohol dehydrogenase from Thermococcus strain AN1. Methods Enzymol. 2001;331:201–7. doi:10.1016/S0076-6879(01)31058-3.
Takahata Y, Nishijima M, Hoaki T, Maruyama T. Thermotoga petrophila sp. nov. and Thermotoga naphthophila sp. nov., two hyperthermophilic bacteria from the Kubiki oil reservoir in Niigata, Japan. Int J Syst Evol Microbiol. 2001;51:1901–9.
Stewart LC, Jung JH, Kim YT, Kwon SW, Park CS, Holden JF. Methanocaldococcus bathoardescens sp. Nov., a hyperthermophilic methanogen isolated from a volcanically active deep-sea hydrothermal vent. Int J Syst Evol Microbiol. 2015;65:1280–3.
Alain K, Marteinsson VT, Miroshnickenko ML, Bonch-Osmolovskaya E, Prieur D, Birrien J-L. Marinitoga piezophila sp. nov., a rod-shaped, thermo-piezophilic bacterium isolated under high hydrostatic pressure from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol. 2002;52:1331–9.
Hoster F, Danie R, Gottschalk G. Isolation of a new Thermoanaerobacterium thermosaccharolyticum strain (FH1) producing a thermostable dextranase. J Gen Appl Microbiol. 2001;47:187–92.
Patel BKC, Morgan HW, Daniel RM. Fervidobacterium nodosum gen. nov. and spec. nov., a new chemoorganotrophic, caldoactive, anaerobic bacterium. Arch Microbiol. 1985;141:63–9.
Willquist K, Van Niel EWJ. Growth and hydrogen production characteristics of Caldicellulosiruptor saccharolyticus on chemically defined minimal media. Int J Hydrogen Energy. 2012;37:4925–9. doi:10.1016/j.ijhydene.2011.12.055.
Mai V, Lorenz WW, Wiegel J. Transformation of Thermoanaerobacterium sp. strain JW/SL-YS485 with plasmid pIKM1 conferring kanamycin resistance. FEMS Microbiol Lett. 1997;148:163–7.
Authors’ contributions
TZ, and DGO conceived the study. HB built the plasmids and strains in this study, and performed preliminary fermentation experiments under supervision of TZ. JC carried out fermentation studies, performed phylogenetic analysis and generated all tables and figures. TZ and JC drafted the manuscript, together with DGO and LRL, who also supervised this study.
Acknowledgements
This work is supported by the BioEnergy Science Center (BESC), a US Department of Energy (DOE) Bioenergy Research Center supported by the Office of Biology and Environmental Research in the DOE Office of Science. Notice: this manuscript has been authored by Dartmouth College under Contract No. DE-AC05-00OR22725 with US Department of Energy. The US Government and the publisher, by accepting the article for publication, acknowledges that the US Government retains a non-exclusive, paid-up, irrevocable worldwide license to publish or reproduce the published form of this manuscript or allow others to do so, for US Government purposes.
Competing interests
Lee R. Lynd is a founder of the Enchi Corporation, which has a financial interest in Clostridium thermocellum.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zheng, T., Cui, J., Bae, H.R. et al. Expression of adhA from different organisms in Clostridium thermocellum . Biotechnol Biofuels 10, 251 (2017). https://doi.org/10.1186/s13068-017-0940-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13068-017-0940-8