Abstract
Background
Ventilator-associated pneumonia (VAP) is a common and serious problem in intensive care units (ICUs). Several studies have suggested that the Gram stain of endotracheal aspirates is a useful method for accurately diagnosing VAP. However, the usefulness of the Gram stain in predicting which microorganisms cause VAP has not been established. The purpose of this study was to evaluate whether a Gram stain of endotracheal aspirates could be used to determine appropriate initial antimicrobial therapy for VAP.
Methods
Data on consecutive episodes of microbiologically confirmed VAP were collected from February 2013 to February 2016 in the ICU of a tertiary care hospital in Japan. We constructed two hypothetical empirical antimicrobial treatment algorithms for VAP: a guidelines-based algorithm (GLBA) based on the recommendations of the American Thoracic Society-Infectious Diseases Society of America (ATS-IDSA) guidelines and a Gram stain-based algorithm (GSBA) which limited the choice of initial antimicrobials according to the results of bedside Gram stains. The GLBA and the GSBA were retrospectively reviewed for each VAP episode. The initial coverage rates and the selection of broad-spectrum antimicrobial agents were compared between the two algorithms.
Results
During the study period, 219 suspected VAP episodes were observed and 131 episodes were assessed for analysis. Appropriate antimicrobial coverage rates were not significantly different between the two algorithms (GLBA 95.4% versus GSBA 92.4%; p = 0.134). The number of episodes for which antimethicillin-resistant Staphylococcus aureus agents were selected as an initial treatment was larger in the GLBA than in the GSBA (71.0% versus 31.3%; p < 0.001), as were the number of episodes for which antipseudomonal agents were recommended as an initial treatment (70.2% versus 51.9%; p < 0.001).
Conclusions
Antimicrobial treatment based on Gram stain results may restrict the administration of broad-spectrum antimicrobial agents without increasing the risk of treatment failure.
Trial registration
UMIN-CTR, UMIN000026457. Registered 8 March 2017 (retrospectively registered).
Similar content being viewed by others
Background
Ventilator-associated pneumonia (VAP) is a serious healthcare-associated infection, common in intensive care units (ICUs). It complicates the medical course of 10–20% of mechanically ventilated patients and results in an estimated attributable mortality of 15–50% [1,2,3]. The selection of initial antimicrobial therapy for VAP is important since inappropriate initial antimicrobial treatment is associated with higher mortality and longer ICU stay [4,5,6,7,8]. Therefore, the 2005 American Thoracic Society-Infectious Diseases Society of America (ATS-IDSA) guidelines on healthcare-associated pneumonia recommend that empirical treatment of patients at risk of multi-resistant organisms should be broad spectrum to cover both multidrug-resistant (MDR) Gram-negative pathogens and methicillin-resistant Staphylococcus aureus (MRSA) [9].
Although early broad-spectrum treatment helps to ensure that infections are treated effectively, overuse of broad-spectrum antimicrobial agents is driving antimicrobial resistance [10, 11]. As VAP caused by multi-resistant Gram-negative bacilli results in significantly higher mortality than VAP caused by other pathogens [12, 13], preventing the emergence of resistant pathogens in the ICU is essential. One of the strategies which could potentially curtail the development of antimicrobial resistance is to use narrow-spectrum antimicrobial agents. In fact, national programmes aimed at combining judicious overall use of antimicrobial agents with narrow-spectrum agents have been associated with reductions in antimicrobial resistance [14, 15]. However, there are no well-established methods to safely restrict use of broad-spectrum antimicrobial agents for VAP.
While a culture of endotracheal aspirate is the gold standard for confirming the causative organisms of VAP, it takes at least 48–72 h to obtain the results of antimicrobial susceptibility tests. On the other hand, a Gram stain of respiratory specimens can provide immediate information about predicted pathogenic bacteria. Several studies have demonstrated the effectiveness of the Gram stain of endotracheal aspirate for diagnosing VAP [16,17,18,19]. However, its effectiveness in predicting causative organisms and guiding appropriate initial antimicrobial therapy has not been well established.
In our ICU, Gram stains of endotracheal aspirates are routinely performed by attending physicians to help diagnose VAP. However, the results of Gram stains have not been used to guide initial antimicrobial therapy. Thus, this study aimed to determine whether the Gram stain of endotracheal aspirate was a reliable guide for selecting appropriate antimicrobial therapy for VAP.
Methods
Study population
This was a retrospective study conducted from February 2013 to February 2016 in the 18-bed ICU of a tertiary care hospital in Japan. Every patient admitted to our ICU during the study period was considered for the study if receiving mechanical ventilation for more than 2 days. Patients who were treated for VAP were eligible for this study. In order to select patients with a likely diagnosis of VAP, we included only those patients who had a modified clinical pulmonary infection score (CPIS) of 5 or more [20] and from whom semi-quantitative growth of a respiratory pathogen using a respiratory sample was estimated to be at least 1+. Patients with missing data relating to Gram stain or sputum culture were excluded. Only the first episode of suspected VAP was included (Fig. 1).
Management
Methods for preventing VAP were based on the guidelines published by the Society for Healthcare and Epidemiology of America (SHEA) [21]: we managed patients without sedation whenever possible; we assessed readiness to extubate daily; we changed the ventilator circuit only if visibly soiled or malfunctioning; and we elevated the head of the bed to 30–45° if not contraindicated.
In our institute, Gram stain of endotracheal aspirate was performed on every patient with suspected VAP and the respiratory sample was sent to a laboratory for sputum culture. Gram stain was performed by the Favor method: heat-fixed smears on slides were flooded with 0.2% Victoria blue for 30 s and then washed with tap water; smears were decolourized with 2% Picric Acid ethanol; cells were counterstained with 0.004% Fuchsin for 30 s and then washed with tap water. Residents trained in this method performed a Gram stain as soon as possible after the respiratory sample was collected. Attending physicians evaluated physical findings, chest radiographs, and the results of Gram stains in reaching a diagnosis of VAP and determined an initial treatment strategy. Broncho-alveolar lavage (BAL) is not systematically performed at the study ICU.
Development of the algorithms
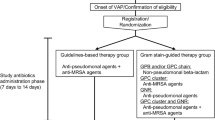
To evaluate whether the Gram stain of endotracheal aspirate was a reliable guide for the selection of antimicrobial therapy for VAP we constructed two hypothetical empirical antimicrobial treatment algorithms: a guidelines-based algorithm (GLBA) and a Gram stain-based algorithm (GSBA).
In the GLBA (Fig. 2a), patients were initially assessed to determine whether they were in septic shock or not. A combination of a carbapenem and an anti-MRSA agent was selected to treat patients in septic shock. If patients did not present with septic shock, initial antibiotics were selected on the basis of the clinical risk factors for MDR pathogens according to ATS-IDSA guidelines [9]. The risk factors were defined as antimicrobial therapy in the preceding 90 days, a hospital stay of 96 h or more, chronic dialysis, immunosuppressive disease or therapy, nursing home admission, or colonizing MDR pathogens from surveillance cultures of endotracheal aspirate collected once a week. If patients had no MDR risk factors, a non-pseudomonal beta-lactam antibiotic was selected as an initial therapy. If patients had at least 1 of the MDR risks, the combination of an anti-pseudomonal agent and an anti-MRSA agent was selected. Finally, if drug-resistant pathogens were isolated from respiratory samples collected before the onset of VAP, we escalated an initial treatment selection process to cover them.
In the GSBA (Fig. 2b), the combination of a carbapenem and an anti-MRSA agent was selected for patients in septic shock, and the risk of MDR pathogens was assessed for patients not in septic shock using the same procedure as with the GLBA. If patients had no MDR risk factors, a non-pseudomonal beta-lactam antibiotic was selected as an initial therapy. If they had at least 1 of the MDR risks, results of bedside Gram stain of endotracheal aspirate were used to guide selection of initial treatment. The results of bedside Gram stains were categorized as Gram-positive cocci (GPC) chains, Gram-positive bacilli (GPB), GPC clusters, Gram-negative rods (GNR), or a combination of these. A non-pseudomonal beta-lactam antibiotic was selected for patients when the Gram stain of the endotracheal aspirate showed only GPC chains and/or GPB. An anti-MRSA agent was selected for patients when the Gram stain results showed GPC clusters without GNR. An anti-pseudomonal agent was selected for patients when the Gram stain results showed GNR without GPC clusters. The combination of an anti-pseudomonal agent and an anti-MRSA agent was selected for patients when the Gram stain results showed both GPC clusters and GNR. We escalated an initial treatment selection process to cover pathogens isolated from respiratory samples collected before the onset of VAP if the Gram stain results suggested their involvement.
In both algorithms, specific antimicrobial agents were selected according to previously recorded antimicrobial resistance patterns in our ICU.
Outcome measures
Both the GLBA and the GSBA were hypothetically applied to the same VAP episodes and the antimicrobial agents proposed by each algorithm were compared retrospectively. Therapy was considered appropriate when all pathogens involved in the VAP episode were covered by antimicrobial agents. We defined the primary endpoint as the coverage rates of initial antimicrobial therapies. Secondary endpoints were set as the selected rates of anti-pseudomonal agents and anti-MRSA agents.
Statistical analysis
Continuous variables were described as mean (± standard deviation) or median (interquartile range) for normal or non-normal distributions, respectively. To compare paired proportions, McNemar’s test for related samples was used. Differences in distribution were checked using the Wilcoxon signed-rank test.
All hypotheses for statistical tests were two-sided, and p values <0.05 were considered to indicate statistical significance. Statistical analyses were performed with R software (version 3.0.2; R Development Core Team) for Windows®.
Results
Patients
Figure 1 shows a patient diagram. There were 219 consecutive suspected VAP episodes during the study period, and 131 of them were used for analysis. The baseline characteristics of the study population are shown in Table 1. The median age of the patients was 67 years (range 48–80 years), and 87 (66.4%) were male. Clinical risk factors for MDR pathogens were present in 89 (67.9%) of the VAP episodes as follows: antimicrobial therapy in the preceding 90 days in 66 (50.4%), a hospital stay of 5 days or more in 70 (53.4%), chronic dialysis in 7 (5.3%), immunosuppressive disease or therapy in 6 (4.6%), nursing home admission in 7 (5.3%), and colonizing MDR pathogens from surveillance cultures of endotracheal aspirate in 17 (13.0%). The median Acute Physiology and Chronic Health Evaluation (APACHE) II score was 25 (range 19–30).
Appropriateness and spectrum of antimicrobial therapy
Antimicrobial choices proposed by the GLBA and the GSBA were appropriate in 95.4% and 92.4% of cases, respectively. McNemar’s test showed no significant difference in adequacy for the different strategies (p = 0.134). The GSBA proposed anti-MRSA agents in significantly fewer episodes than the GLBA (41 (31.3%) versus 93 (71.0%), respectively; p < 0.001). The GSBA also recommended antipseudomonal agents in significantly fewer episodes than the GLBA (68 (51.9%) versus 92 (70.2%), respectively; p < 0.001) (Fig. 3).
Twenty-two MDR pathogens defined by the Centers for Disease Control and Prevention criteria were isolated: two extended spectrum beta-lactamase (ESBL)-positive enterobacteriaceae and 20 MRSA. In the GLBA, there were six inappropriate antimicrobial choices (4.6%): in five patients with no MDR risk factors, ampicillin sulbactam-resistant GNR was isolated (3.8%); in the other patient with at least one MDR risk factor, one ESBL-positive E. coli (0.8%) was isolated. Using the GSBA, 10 inappropriate antimicrobial choices were observed (7.6%): in five patients without any MDR risk factors, non-fermenter pathogens and MRSA were isolated (3 (2.3%) and 2 (1.5%), respectively); in the other five patients with at least one MDR risk factor, one ESBL-positive E. coli (0.8%) and one drug-resistant Corynebacterium sp. (0.8%) were isolated. Inappropriate antimicrobial choices due to misidentification of pathogens in the Gram-stained aspirate were observed in only three cases (2.3%), misidentification of GNR accounted for one case, and the other two were caused by misidentification of GPC clusters.
Pathogens and Gram stain
From the 131 endotracheal aspirates that were Gram stained and cultured, 209 pathogens were isolated. Details of pathogens isolated are shown Table 2.
GPC clusters were identified on Gram stain in 64 patients, and S. aureus was cultured in 50 (78.1%) of these samples. Of the 67 patients from whom GPC clusters were not identified on Gram stain, S. aureus was cultured in nine samples (13.4%). The false-negative rates of GPC clusters were 15.3% (95% confidence interval (CI) 0.072–0.270) (Table 3). However, Gram-negative pathogens were cultured in 71 samples (74.7%) taken from 95 patients from whom GNR was detected on Gram stain. Gram-negative pathogens were cultured in four samples (6.7%) from 60 patients from whom GNR were not detected on Gram stain. The false-negative rates of GNR were 5.3% (95% CI 0.015–0.131) (Table 4).
Discussion
The present study focused on the effectiveness of Gram stain to guide initial treatment selection in patients with VAP. The GSBA had a satisfactorily high coverage rate and suggested significantly lesser use of anti-MRSA agents and anti-pseudomonal agents than did the GLBA. Thus, the main findings were that the treatment algorithm constructed on the basis of Gram stain results could reduce the use of broad-spectrum antimicrobials without increasing the risk of treatment failure.
The guidelines for management of adult VAP published by the Infectious Diseases Society of America (IDSA) [22] have emphasized the importance of reducing the unnecessary use of broad-spectrum antimicrobials in order to minimize patient harm and reduce the development of antimicrobial resistance. Gram stain is a potentially useful tool to limit empirical broad-spectrum antimicrobial therapy in patients with VAP because it can provide information on predicted causative organisms promptly. However, whether Gram stain is accurate enough for the use of broad-spectrum antimicrobials to be safely restricted is still controversial. To the best of our knowledge, this study provides the first evidence that Gram stain of endotracheal aspirates may reduce the use of broad-spectrum antimicrobials without lowering appropriate coverage rates compared to a guidelines-based method.
In this study, the false negative rates of Gram stain were low both for GPC clusters (15.3%, 95% CI 0.072–0.270) and GNR (5.3%, 95% CI 0.015–0.131). The results of our study are consistent with previous studies that have reported high sensitivity and low false-negative rates for Gram stain used to identify Staphylococcus aureus [23,24,25]. On the other hand, some studies have reported only a fair correlation between Gram stain and culture [16, 26]. In these studies, some of the patients did not actually have VAP and might have had low-level colonization. Besides, the timing of the collected endotracheal aspirate was not clearly reported. In the present study, we only included confirmed VAP patients with modified CPIS of 5 or more and patients who had a high level of colonization—at least 1+ semiquantitative growth. In addition, all endotracheal aspirates were collected before the administration of the antimicrobials. These differences might have led to the considerably better correlation between Gram stain and culture in the current study. Moreover, a misidentification of GPC clusters from Gram stain did not necessarily result in insufficient coverage as selected antibiotics could cover methicillin-susceptible S. aureus. Similarly, misidentification of GNR was not detrimental unless non-fermenters or MDR pathogens were cultured. Therefore, we observed quite high appropriate coverage rates using the GSBA.
Several limitations have to be addressed. First, as this was a retrospective analysis of two hypothetical algorithms, the performance of both algorithms in reality could be different from the observed results. However, the findings from the Gram stains were independently confirmed by the final results from cultures of endotracheal aspirates. The superiority of the GSBA over the GLBA was derived from the high concordance rates between the results of Gram stain and sputum culture, indicating that, despite the retrospective study design, the Gram-stain diagnostic method would have high efficacy in clinical practice. Second, as the risk of inappropriate antimicrobial choices due to false-negative Gram stain results depends on the resistance rates of causative organisms, the coverage rates of GSBA alter according to local susceptibility in each ICU. Third, we could not evaluate whether patient outcomes from the GSBA were superior to the GLBA. Thus, a prospective study must be conducted to assess patient outcomes from antimicrobial treatment based on Gram stain results.
Conclusions
Compared with guidelines-based treatment, bedside Gram stains may reduce the use of broad-spectrum antimicrobials for patients with VAP without lowering appropriate coverage rates.
Abbreviations
- APACHE:
-
Acute Physiology and Chronic Health Evaluation
- ATS-IDSA:
-
American Thoracic Society-Infectious Diseases Society of America
- BAL:
-
Broncho-alveolar lavage
- CI:
-
Confidence interval
- CPIS:
-
Clinical pulmonary infection score
- ESBL:
-
Extended spectrum beta-lactamase
- GLBA:
-
Guidelines-based algorithm
- GNR:
-
Gram-negative rods
- GPB:
-
Gram-positive bacilli
- GPC:
-
Gram-positive cocci
- GSBA:
-
Gram stain-based algorithm
- ICU:
-
Intensive care unit
- IDSA:
-
Infectious Diseases Society of America
- MDR:
-
Multidrug-resistant
- MRSA:
-
Methicillin-resistant Staphylococcus aureus
- SHEA:
-
Society for Healthcare and Epidemiology of America
- VAP:
-
Ventilator-associated pneumonia
References
Safdar N, Dezfulian C, Collard HR, Saint S. Clinical and economic consequences of ventilator-associated pneumonia: a systematic review. Crit Care Med. 2005;33:2184–93.
Hortal J, Muñoz P, Cuerpo G, Litvan H, Rosseel PM, Bouza E. Ventilator-associated pneumonia in patients undergoing major heart surgery: an incidence study in Europe. Crit Care. 2009;13:R80.
Forel JM, Voillet F, Pulina D, Gacouin A, Perrin G, Barrau K, et al. Ventilator-associated pneumonia and ICU mortality in severe ARDS patients ventilated according to a lung-protective strategy. Crit Care. 2012;16:R65.
Dupont H, Mentec H, Bleichner G. Impact of appropriateness of initial antibiotic therapy on the outcome of ventilator-associated pneumonia. Intensive Care Med. 2001;27:355–62.
Luna CM, Vujacich P, Niederman MS, Vay C, Gherardi C, Matera J, Jolly EC. Impact of BAL data on the therapy and outcome of ventilator-associated pneumonia. Chest. 1997;111:676–85.
Kollef MH, Ward S. The influence of mini-BAL cultures on patient outcomes: implications for the antibiotic management of ventilator-associated pneumonia. Chest. 1998;113:412–20.
Kollef MH, Sherman G, Ward S, Fraser VJ. Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients. Chest. 1999;115:462–74.
Trouillet JL, Chastre J, Vuagnat A, Joly-Guillou ML, Combaux D, Dombret MC, et al. Ventilator-associated pneumonia caused by potentially drug-resistant bacteria. Am J Respir Crit Care Med. 1998;157:531–9.
American Thoracic Society, Infectious Diseases Society of America. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med. 2005;171:388–416.
Shlaes DM, Gerding DN, John Jr JF, Craig WA, Bornstein DL, Duncan RA, et al. Society for Healthcare Epidemiology of America and Infectious Diseases Society of America Joint Committee on the Prevention of Antimicrobial Resistance: guidelines for the prevention of antimicrobial resistance in hospitals. Infect Control Hosp Epidemiol. 1997;18:275–91.
Kaki R, Elligsen M, Walker S, Simor A, Palmay L, Daneman N. Impact of antimicrobial stewardship in critical care: a systematic review. J Antimicrob Chemother. 2011;66:1223–30.
Kollef MH, Silver P, Murphy DM, Trovillion E. The effect of late-onset ventilator-associated pneumonia in determining patient mortality. Chest. 1995;108:1655–62.
Depuydt PO, Vandijck DM, Bekaert M, Decruyenaere JM, Blot SI, Vogelaers DP, et al. Determinants and impact of multidrug antibiotic resistance in pathogens causing ventilator-associated-pneumonia. Crit Care. 2008;12:R142.
Mölstad S, Cars O. Major change in the use of antibiotics following a national programme: Swedish Strategic Programme for the Rational Use of Antimicrobial Agents and Surveillance of Resistance (STRAMA). Scand J Infect Dis. 1999;31:191–5.
Seppala H, Klaukka T, Vuopio-Varkila J, Muotiala A, Helenius H, Lager K, Huovinen P. The effect of changes in the consumption of macrolide antibiotics on erythromycin resistance in group A streptococci in Finland. N Engl J Med. 1997;337:441–6.
O'Horo JC, Thompson D, Safdar N. Is the gram stain useful in the microbiologic diagnosis of VAP? A meta-analysis. Clin Infect Dis. 2012;55:551–61.
Sole-Violan J, Rodriguez de Castro F, Rey A, Martin-Gonzalez JC, Cabrera-Navarro P. Usefulness of microscopic examination of intracellular organisms in lavage fluid in ventilator-associated pneumonia. Chest. 1994;106:889–94.
Allaouchiche B, Jaumain H, Chassard D, Bouletreau P. Gram stain of bronchoalveolar lavage fluid in the early diagnosis of ventilator-associated pneumonia. Br J Anaesth. 1999;83:845–9.
Brun-Buisson C, Fartoukh M, Lechapt E, Honoré S, Zahar JR, Cerf C, et al. Contribution of blinded, protected quantitative specimens to the diagnostic and therapeutic management of ventilator-associated pneumonia. Chest. 2005;128:533–44.
Luna CM, Blanzaco D, Niederman MS, Matarucco W, Baredes NC, Desmery P, et al. Resolution of ventilator-associated pneumonia: prospective evaluation of the clinical pulmonary infection score as an early clinical predictor of outcome. Crit Care Med. 2003;31:676–82.
Klompas M, Branson R, Eichenwald EC, Greene LR, Howell MD, Lee G, et al. Strategies to prevent ventilator-associated pneumonia in acute care hospitals: 2014 update. Infect Control Hosp Epidemiol. 2014;35 Suppl 2:133–54.
Kalil AC, Metersky ML, Klompas M, Muscedere J, Sweeney DA, Palmer LB, et al. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis. 2016. doi:10.1093/cid/ciw353.
Seligman R, Seligman BG, Konkewicz L, Pires dos Santos R. Accuracy of tracheal aspirate gram stain in predicting Staphylococcus aureus infection in ventilator-associated pneumonia. BMC Anesthesiol. 2015;15:19.
Blot F, Raynard B, Chachaty E, Tancrède C, Antoun S, Nitenberg G. Value of gram stain examination of lower respiratory tract secretions for early diagnosis of nosocomial pneumonia. Am J Respir Crit Care Med. 2000;162:1731–7.
Gottesman T, Yossepowitch O, Lerner E, Schwartz-Harari O, Soroksky A, Yekutieli D. The accuracy of Gram stain of respiratory specimens in excluding Staphylococcus aureus in ventilator-associated pneumonia. J Crit Care. 2014;29:739–42.
Tetenta S, Metersky ML. Tracheal aspirate Gram stain has limited sensitivity and specificity for detecting Staphylococcus aureus. Respirology. 2011;16:86–9.
Acknowledgements
The authors would like to thank John Martin for his linguistic assistance with the manuscript.
Funding
No financial support was received for the performance of this study.
Availability of data and materials
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
JY conceived, designed, and coordinated the study, performed data acquisition and analyses, and drafted the manuscript. TK participated in data collection and interpretation, and helped to draft the manuscript. TH performed statistical analysis. KY, AM, and NN contributed to data interpretation and critically reviewed the manuscript. SF exerted a major impact on the interpretation of data and critical appraisal of the manuscript. All authors have read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study followed the principles of the Declaration of Helsinki and was approved by the institutional review board of Osaka General Medical Center (reference number: 28-C0107). The board waived the need for informed consent because this was a retrospective chart review study.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yoshimura, J., Kinoshita, T., Yamakawa, K. et al. Impact of Gram stain results on initial treatment selection in patients with ventilator-associated pneumonia: a retrospective analysis of two treatment algorithms. Crit Care 21, 156 (2017). https://doi.org/10.1186/s13054-017-1747-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13054-017-1747-5