Abstract
Background
Breast cancer develops as a result of multiple gene mutations in combination with environmental risk factors. Causative variants in genes such as BRCA1 and/or BRCA2 have been shown to account for hereditary nature of certain breast cancers. However,other genes, such as ATM, PALB2, BRIP1, CHEK, BARD1, while lower in frequency, may also increase breast cancer risk. There are few studies examining the role of these causative variants. Our study aimed to examine the clinical and genetic characterization of hereditary breast cancer in a Chinese population.
Methods
We tested a panel of 27 genes implicated in breast cancer risk in 240 participants using Next-Generation Sequencing. The prevalence of genetic causative variants was determined and the association between causative variants and clinico-pathological characteristics was analyzed.
Results
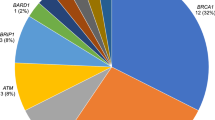
Causative variant rate was 19.2% in the breast cancer (case) group and 12.5% in the high-risk group. In the case group 2.5% of patients carried BRCA1 causative variant, 7.5% BRCA2 variants, 1.7% patients had MUTYH, CHEK or PALB2 variants, and 0.8% patients carried ATM, BARD1, NBN, RAD51C or TP53 variants. In the high-risk group 5.8% women carried MUTYH causative variants, 2.5% had causative variants in ATM, 1.7% patients had variants in BRCA2 and 0.8% in BARD1, BRIP1 or CDH1. There was no significant difference in the presence of causative variants among clinical stages of breast cancer, tumor size and lymph nodes status. However, eight of the 12 BRCA1/2 causative variants were found in the TNBC group.
Conclusions
We found increased genetic causative variants in the familial breast cancer group and in high-risk women with a family history of breast cancer. However, the variant MUTYH c.892-2A > G may not be directly associated with hereditary breast carcinoma.
Similar content being viewed by others
Background
Breast cancer is a common malignancy among women, with an estimated annual rate of incidence increasing by 2–3% in China, especially in metropolitan areas [1]. It is known that while the majority of breast cancers are sporadic in origin, an appreciable fraction result from inherited causative variants [2, 3] . Cancer is caused by the cumulative effects of mutations in multiple genes, in combination with environmental factors. It has been suggested that reproductive and hormonal factors, such as nulliparity, increased age at first live birth, and limited breast feeding are associated with a modest increase in the risk of breast cancer in Western countries [4, 5]. Breast cancer susceptibility genes BRCA1 and BRCA2 causative variant account for only 10–20% of breast cancers with a known family history [6]. The prevalence of hereditary breast cancers is approximately 11.8% in China [7], suggesting that other genes may play an important role in increasing the susceptibility to breast cancer, albeit at a markedly lower frequency and penetrance. For example, women with inherited causative variant in the Fanconi anemia genes BRIP1 and PALB2 have a 20–50% lifetime risk of breast cancer [8, 9]. Multiple studies have also demonstrated that genes such as ATM [10,11,12] and CHEK2 [13,14,15,16] are associated with increased breast cancer risk. In addition, inherited causative variants in TP53, PTEN, STK11, and CDH1 are associated with a moderate to very high-risk of developing breast cancer [17,18,19,20].
Although studies have demonstrated the clinical benefit of multiple-gene sequencing for the assessment of patients with high-risk hereditary cancer [21, 22], little information is currently available regarding the value of multiple-gene sequencing for the assessment of the risk of hereditary breast cancer in China. The goal of this study was to identify the variant spectrum for the clinical and genetic characterization of familial breast cancer in a Chinese population. Twenty-seven breast cancer susceptibility genes (Additional file 1: Table S1), selected through a database (HGMD: Human Gene Mutation Database, NCBI ClinVar database) and published research articles, were tested by Next-Generation Sequencing (NGS).
Methods
Patients and samples
In total, 240 participants, including 120 patients with breast cancer and 120 high-risk women with first- or second-degree relative(s) suffering from breast cancer were recruited from Shenzhen Second People’s Hospital of China during a two year period (2014–2016). The rate of susceptibility gene causative variants in East Asian population in 1000 Genomes database was used as a control. The clinical breast cancer diagnosis and classification criteria were in accordance with the World Health Organization criteria. Written informed consent was obtained from patients and healthy high-risk women. The study was approved by a local ethics committee. Two hundred and forty peripheral blood samples were collected and referred for genetic testing to the BGI research Department (Shenzhen, China).
Sample treatment, next-generation sequencing and variants calling
DNA was extracted from participants’ peripheral blood samples using a Qiagen DNA blood mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations. Qubit Fluorometer (Life Technologies) and agarose gel electrophoresis were used to determine DNA concentration and purity. Genomic DNAs were randomly fragmented to 200-300 bp by Covaris E210 (Massachusetts, USA) and treated as follows: end-repair, A-tailing and adapter ligation, and PCR amplification. PCR products were captured by the same BGI chip in the Blackbird platform. Their frequency was determined by quantitative PCR, and the segments were pooled for sequencing on the Hiseq 2500 (Illumina) according to the manufacturer’s protocols. Over 0.6 GB data was generated per sample with approximately 200X depth and over 99% coverage of the target region. Variants were detected using Small Variant Assembler Methods (http://www.completegenomics.com/documents/Small_Variant_Assembler_Methods.pdf) which is available on the official website of Complete Genomics. Then, variants were filtered according to their read support, assemble quality and reference allele repeat status. Sequences generated by high-throughput sequencing platforms were filtered by SOAPnuke1.5.0 with standard augmentation, and then assembled by BWA 0.7.12 using MEM. Sam Tools 1.2 was used to convert file format into BAM. Base quality was recalibrated by GATK 3.4. Duplications were removed by Picard Mark Duplicates 1.138. Local realignment of reads around insertion/deletion was performed and variants were called by insertion/deletion Realigner and Haplotype Caller in GATK 3.4. Variants were further filtered by quality depth, strand bias, mapping quality and reads position.
Variant classification
In accordance with the American College of Medical Genetics (ACMG) recommendations for the interpretation of sequence, variants were classified into pathogenic, likely pathogenic, variant of uncertain significance (VUS), likely benign, and benign variant. Variants were classified as pathogenic if they conferred truncations, or initiation codons, affected splicing or if they have been reported in the central mutation database (HGMD, ClinVar), or in published literature, and demonstrated to be causative of the disorder in a particular disease with no conflicting results. Variants were classified as VUS if they fulfilled the following three criteria at the same time: 1) missense, non-frame shift or intronic (exon-intron boundaries ±10 bp) variants, and 2) allele frequency in the 1000 Genomes Study and 101 BGI normal Chinese genomes study are both less than 0.03, and 3) variants were not uniformly identified as benign/likely benign in ClinVar. The rest of variants were identified as benign. In addition, every pathogenic variant detected by next-generation sequencing was confirmed by conventional PCR-Sanger sequencing. Twenty-seven genes examined in this study (Additional file 1: Table S1) were selected through database or published articles about known mutations in hereditary breast cancer.
Statistics
Statistical tests were carried out using SPSS 20.0 (IBM, Armonk, NY), applying chi-square or Fisher’s exact tests when required to analyze categorical data. A p values less than 0.05 was considered as statistically significant.
Results
Characteristics of the study population
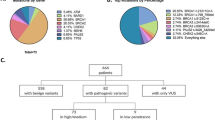
We recruited for this trial 120 patients diagnosed with breast cancer and 120 high-risk women who had first-degree relatives affected by breast cancer. Table 1 summarizes the risk factor data of the study population reflecting the epidemiology of breast cancer. The median age at blood sample collection was 46 years (range from 25 to 81 years) in the breast cancer group and the median age was 37 years in the high-risk group. There were no statistically significant differences in body mass index (BMI), age at menarche, and breast-feeding history. However, there were statistically significant differences between the two groups in parity and abortion rates. In this study 77.5% of patients had no history of childbearing and 41.7% of patients had a history of abortion, which may confer a high-risk of breast cancer in Chinese individuals.
Prevalence of panel-gene causative variants in the two groups
In order to explore the presence of predisposing genetic factors for the development of breast cancer, all participants were subjected to a multiple-gene panel sequencing and variant analysis. The presence of 27 causative variants (Additional file 1: Table S1) associated with an increased susceptibility to breast cancer was tested in this panel using NGS. As showed in Table 2, the ratio of variants in the breast cancer group was 19.2% (23/120) and 12.5% (15/120) in the high-risk group. Twelve predisposing causative variants in 27 panel-genes were identified in this study. Three (2.5%) in BRCA1, nine (7.5%) in BRCA2, two (1.7%) each in MUTYH, CHEK and PALB2, one (0.8%) each in ATM, BARD1, NBN, RAD51C, TP53 were identified in the breast cancer group, while seven (5.8%) in MUTYH, three (2.5%) in ATM, two (1.7%) in BRCA2, one (0.8%) each in BARD1, BRIP1 and CDH1 were identified in the high-risk group. There were no causative variants found in other genes examined.
All germline changes revealed by panel sequencing were termed germ line causative variants by the 5-tier rating system. We have excluded “likely benign”, “benign” variants and VUS in the paper, and have listed “pathogenic”, “likely pathogenic,” changes in Tables 3 and 4. Detailed information regarding causative variants in the breast cancer group and the high-risk group (women with a family history of breast cancer) is listed in Tables 3 and in Table 4. Genetic causative variants identified were heterozygous mutations, and most were frameshift deletions. We did not include healthy women with no known history of familial breast cancer in our study, however frequencies of gene causative variants that we identified were examined in healthy population by surveying available databases: http://www.internationalgenome.org/ and http://www.ncbi.nlm.nih.gov/projects/SNP/. We found that the frequencies of these variants were zero in East Asian population in 1000G_ALL (the frequency of this causative variants in all populations of the human international genome). However, we detected MUTYH gene variants (Intron10, c.892-2A > G) at a rate of 2.77% https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=77542170 in East Asian healthy individuals.
Association between genetic causative variants and clinicopathological characteristics
Gene causative variants prevalence was 69.6% (16/23) in patients with invasive ductal carcinoma (IDC), 4.3% (1/23) patients with ductal carcinoma in situ (DCIS) and 26.1% (6/23) with an unknown histological type (Table 5). There was no significant difference in the presence of variants between clinical stages of breast cancer (Pearson’s Chi-squared test p = 0.537). Although some patients were lost to follow-up, our data suggest that similar causative variants were found in patients regardless of tumor size and lymph nodes status.
When analyzed, based on the molecular subtype of breast cancer, the genetic causative variant ratio was 43.5% in patients with triple negative breast cancer (TNBC), 39.1% in patients with non-TNBC, and 17.4% in patients with undetermined molecular subtype (p = 0.001) (Table 5). Eight of the 12 BRCA1/2 causative variants were found in the TNBC group. The other two gene variants in the TNBC group were BARD1 and RAD51.
Discussion
In this clinical study, we examined 27 genes associated with an increased susceptibility to breast cancer (Tables 2, 3 and 4) in patients with breast cancer and in high-risk participants with a family history of breast cancer. In addition to BRCA1/2, genes with an established role in breast cancer, other predisposing genes such as CHEK and PALB2 were evaluated for a possible association with the risk of breast cancer, although their frequency and penetrance was significantly lower. We found causative variants in 12 of the 27 genes examined in the participants (Table 2).
There appeared to be considerable discrepancies in the causative variant rates of BRCA1 and BRCA2 in breast cancer patients in different areas of China. Song [23] reported that the variant ratio of BRCA1 and BRCA2 in Shanghai was 11.4% and 2.9%, respectively, whereas in our study the variant ratio of BRCA1 and BRCA2 in breast cancer patients were 2.5% and 7.5%, respectively (Table 2). The main reason for lower causative variant rates of BRCA1 and higher variant rates of BRCA2 in our study may be the different detection methods used in the studies. PCR-SSCP analysis, examining only four “hot areas” in BRCA1/2 was used in the Song’s study, while whole exon NGS of BRCAs was used in our study. In addition, geographical differences are likely to contribute to discrepancies between results. The participants in the Song’s study mainly were recruited from Eastern and Northern China, while the subjects in our study were largely from Southern and Central China.
We found a relatively high variant rate (4.2%, 5/120) of MUTYH c.892-2A > G in the high-risk group, but lower rate (0.8%, 1/120) in the breast cancer group (Table 2). According to the 5-tier rating system in ACMG, this variant is likely pathogenic. However, a correlation between MUTYH variants and breast cancer remains unclear. For example, two other studies suggested a significantly increased breast cancer risk among carriers of the bi-allelic MUTYH variants [24, 25], while other studies showed that germline MUTYH variants are not associated with carcinomas of the breast [26, 27] . In our study, the variant ratio of MUTYH c.892-2A > G in high-risk women with a family history of breast cancer is over 2.77% https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=77542170, the frequency of MUTYH c.892-2A > G in East Asians in 1000G_ALL, but the rate in the breast cancer group is lower. The variant MUTYH c.892-2A > G identified in our study is a heterozygous mutation (Tables 3 and 4). Further, two family pedigrees suggests segregation of this variant (Fig. 1) - the proband did not carry the variant, while their relatives with no BC carried it. Therefore, it is possible that MUTYH c.892-2A > G is a benign variant in the development of BC in East Asians, however we need to enlarge the sample size to confirm this result.
To explore the relationship between gene variants associated with hereditary predisposition and tumor characteristics, we analyzed the association between available pathological and clinical data in breast cancer patients and the presence of gene causative variants. Our results show no statistically significant differences between the presence of gene variants in breast cancer patients and differences in tumor histology, size, clinical stage and lymph node status, however; we found a statistically significant difference in the variant rate in patients with tumors of different molecular type (Table 5). Ten of 22 patients with TNBC were found to harbor gene causative variants. Furthermore, most of TNBC patients (8/10) were found to have BRCA1/2 causative variants. It has been reported that TNBC is common in BRCAs variant carriers [28,29,30,31]. Indeed, the incidence of TNBC is around 70% in BRCA1 mutation carriers [32, 33]. Our data are consistent with this observation, however we need to enlarge the sample size to further confirm this association.
As for the clinical significance of the presence of predisposing variants, different advice may be given to specific groups of patients. Patients carrying these pathogenic variants are considered to be at a high-risk in developing tumor recurrence or secondary cancer according to the NCCN guidelines [9, 34]. However, contralateral mastectomy or oophorectomy for these patients is currently not recommended in China, and asymptomatic women carrying pathogenic variants usually prefer not to undergo preventive surgery. In light of this situation, we suggest that patients with a high-risk of developing breast cancer have a comprehensive physical exam every six months, and we advise them to focus on breast self-examination and maintain a healthy life style.
Conclusion
As the incidence of breast cancer is increasing, it is necessary to carry out more studies to identify susceptibility genes of breast cancer and to establish their frequency. Our results enrich our knowledge of predisposing variants in the population of Southern and Central China, and provide some experimental data for the identification of alternative susceptibility genes, and for the establishment of a clinical model of genetic screening.
However, our study also has some limitations. We did not analyze the relationship between clinicopathological characteristics and gene VUS. More than two hundred VUS were identified in this study, but we have not analyzed them to date. In addition, some patients were lost due to follow-up, which made it difficult to draw conclusions between the association of genetic causative variants and clinicopathological characteristics of patients.
Abbreviations
- DCIS:
-
Ductal carcinoma in situ
- IDC:
-
Invasive ductal carcinoma
- NGS:
-
Next-Generation Sequencing
- TNBC:
-
Triple negative breast cancer
- VUS:
-
Uncertain significance
References
Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, et al. Breast cancer in China. Lancet Oncol 2014;15(7):e279–e289. doi:10.1016/S1470-2045(13)70567-9. PubMed PMID: 24872111.
Collaborative Group on Hormonal Factors in Breast C. Breast cancer and hormonal contraceptives: collaborative reanalysis of individual data on 53 297 women with breast cancer and 100 239 women without breast cancer from 54 epidemiological studies. Lancet 1996;347(9017):1713–1727. PubMed PMID: 8656904.
Breast cancer and hormonal contraceptives: further results. Collaborative Group on Hormonal Factors in Breast Cancer. Contraception. 1996;54(3 Suppl):1S–106S. PubMed PMID: 8899264.
Zhang Q, Liu LY, Wang F, Mu K, Yu ZG. The changes in female physical and childbearing characteristics in China and potential association with risk of breast cancer. BMC Public Health 2012;12:368. doi:10.1186/1471-2458-12-368. PubMed PMID: 22612880; PubMed Central PMCID: PMC3422180.
Bao PP, Shu XO, Gao YT, Zheng Y, Cai H, Deming SL, et al. Association of hormone-related characteristics and breast cancer risk by estrogen receptor/progesterone receptor status in the shanghai breast cancer study. Am J Epidemiol 2011;174(6):661–671. doi:10.1093/aje/kwr145. PubMed PMID: 21768404; PubMed Central PMCID: PMC3166707.
Singletary SE. Rating the risk factors for breast cancer. Ann Surg 2003;237(4):474–482. doi:10.1097/01.SLA.0000059969.64262.87. PubMed PMID: 12677142; PubMed Central PMCID: PMC1514477.
Li WF, Hu Z, Rao NY, Song CG, Zhang B, Cao MZ, et al. The prevalence of BRCA1 and BRCA2 germline mutations in high-risk breast cancer patients of Chinese Han nationality: two recurrent mutations were identified. Breast Cancer Res Treat 2008;110(1):99–109. doi:10.1007/s10549-007-9708-3. PubMed PMID: 17851763.
Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet 2007;39(2):165–167. doi:10.1038/ng1959. PubMed PMID: 17200668; PubMed Central PMCID: PMC2871593.
Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet 2006;38(11):1239–1241. doi:10.1038/ng1902. PubMed PMID: 17033622.
Ahmed M, Rahman N. ATM and breast cancer susceptibility. Oncogene 2006;25(43):5906–5911. doi:10.1038/sj.onc.1209873. PubMed PMID: 16998505.
Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, Ahmed M, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet 2006;38(8):873–875. doi:10.1038/ng1837. PubMed PMID: 16832357.
Thompson D, Duedal S, Kirner J, McGuffog L, Last J, Reiman A, et al. Cancer risks and mortality in heterozygous ATM mutation carriers. J Natl Cancer Inst 2005;97(11):813–822. doi: https://doi.org/10.1093/jnci/dji141. PubMed PMID: 15928302.
Apostolou P, Fostira F. Hereditary breast cancer: the era of new susceptibility genes. Biomed Res Int 2013;2013:747318. doi:10.1155/2013/747318. PubMed PMID: 23586058; PubMed Central PMCID: PMC3618918.
Consortium CBCC-C. CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am J Hum Genet. 2004;74(6):1175–82. doi:10.1086/421251. PubMed PMID: 15122511; PubMed Central PMCID: PMC1182081.
Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet 2002;31(1):55–59. doi:10.1038/ng879. PubMed PMID: 11967536.
Weischer M, Nordestgaard BG, Pharoah P, Bolla MK, Nevanlinna H, Van't Veer LJ, et al. CHEK2*1100delC heterozygosity in women with breast cancer associated with early death, breast cancer-specific death, and increased risk of a second breast cancer. J Clin Oncol 2012;30(35):4308–4316. doi:10.1200/JCO.2012.42.7336. PubMed PMID: 23109706; PubMed Central PMCID: PMC3515767.
Gonzalez KD, Noltner KA, Buzin CH, Gu D, Wen-Fong CY, Nguyen VQ, et al. Beyond li Fraumeni syndrome: clinical characteristics of families with p53 germline mutations. J Clin Oncol 2009;27(8):1250–1256. doi:10.1200/JCO.2008.16.6959. PubMed PMID: 19204208.
Hearle N, Schumacher V, Menko FH, Olschwang S, Boardman LA, Gille JJ, et al. Frequency and spectrum of cancers in the Peutz-Jeghers syndrome. Clin Cancer Res 2006;12(10):3209–3215. doi:10.1158/1078-0432.CCR-06-0083. PubMed PMID: 16707622.
Pharoah PD, Guilford P, Caldas C, International gastric cancer linkage C. Incidence of gastric cancer and breast cancer in CDH1 (E-cadherin) mutation carriers from hereditary diffuse gastric cancer families. Gastroenterology 2001;121(6):1348–1353. PubMed PMID: 11729114.
Tan MH, Mester JL, Ngeow J, Rybicki LA, Orloff MS, Eng C. Lifetime cancer risks in individuals with germline PTEN mutations. Clin Cancer Res 2012;18(2):400–407. doi:10.1158/1078-0432.CCR-11-2283. PubMed PMID: 22252256; PubMed Central PMCID: PMC3261579.
Lin PH, Kuo WH, Huang AC, Lu YS, Lin CH, Kuo SH, et al. Multiple gene sequencing for risk assessment in patients with early-onset or familial breast cancer. Oncotarget 2016;7(7):8310–8320. doi:10.18632/oncotarget.7027. PubMed PMID: 26824983; PubMed Central PMCID: PMC4884994.
Sardanelli F, Podo F. Management of an inherited predisposition to breast cancer. N Engl J Med. 2007;357(16):1663; author reply doi:10.1056/NEJMc072261. PubMed PMID: 17942883.
Song CG, Hu Z, Yuan WT, Di GH, Shen ZZ, Huang W, et al. [BRCA1 and BRCA2 gene mutations of familial breast cancer from shanghai in China]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2006;23(1):27–31. PubMed PMID: 16456781.
Nielsen M, Franken PF, Reinards TH, Weiss MM, Wagner A, van der Klift H, et al. Multiplicity in polyp count and extracolonic manifestations in 40 Dutch patients with MYH associated polyposis coli (MAP). J Med Genet 2005;42(9):e54. doi:10.1136/jmg.2005.033217. PubMed PMID: 16140997; PubMed Central PMCID: PMC1736132.
Vogt S, Jones N, Christian D, Engel C, Nielsen M, Kaufmann A, et al. Expanded extracolonic tumor spectrum in MUTYH-associated polyposis. Gastroenterology. 2009;137(6):1976–85 e1–10. doi:10.1053/j.gastro.2009.08.052. PubMed PMID: 19732775.
Casper M, Acalovschi M, Lammert F, Zimmer V. The MUTYH hotspot mutations p.G396D and p.Y179C do not cause substantial genetic susceptibility to biliary cancer. Familial Cancer 2014;13(2):243–247. doi:10.1007/s10689-014-9699-2. PubMed PMID: 24420788.
Out AA, Wasielewski M, Huijts PE, van Minderhout IJ, Houwing-Duistermaat JJ, Tops CM, et al. MUTYH gene variants and breast cancer in a Dutch case-control study. Breast Cancer Res Treat 2012;134(1):219–227. doi:10.1007/s10549-012-1965-0. PubMed PMID: 22297469; PubMed Central PMCID: PMC3397219.
de Ruijter TC, Veeck J, de Hoon JP, van Engeland M, Tjan-Heijnen VC. Characteristics of triple-negative breast cancer. J Cancer Res Clin Oncol 2011;137(2):183–192. doi:10.1007/s00432-010-0957-x. PubMed PMID: 21069385; PubMed Central PMCID: PMC3018596.
Haffty BG, Yang Q, Reiss M, Kearney T, Higgins SA, Weidhaas J, et al. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol 2006;24(36):5652–5657. doi: https://doi.org/10.1200/JCO.2006.06.5664. PubMed PMID: 17116942.
Ismail-Khan R, Bui MM. A Review of triple-negative breast cancer. Cancer Control 2010;17(3):173–176. PubMed PMID: 20664514.
Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 2003;100(14):8418–8423. doi:10.1073/pnas.0932692100. PubMed PMID: 12829800; PubMed Central PMCID: PMC166244.
Foulkes WD, Stefansson IM, Chappuis PO, Begin LR, Goffin JR, Wong N, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst 2003;95(19):1482–1485. PubMed PMID: 14519755.
Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, et al. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res 2005;11(14):5175–5180. doi:10.1158/1078-0432.CCR-04-2424. PubMed PMID: 16033833.
Daly MB, Pilarski R, Axilbund JE, Berry M, Buys SS, Crawford B, et al. Genetic/familial high-risk assessment: breast and ovarian, version 2.2015. J Natl Compr Cancer Netw 2016;14(2):153–162. PubMed PMID: 26850485.
Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol 2007;25(11):1329–1333. doi:10.1200/JCO.2006.09.1066. PubMed PMID: 17416853; PubMed Central PMCID: PMC2267287.
Pujana MA, Han JD, Starita LM, Stevens KN, Tewari M, Ahn JS, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet 2007;39(11):1338–1349. doi:10.1038/ng.2007.2. PubMed PMID: 17922014.
Win AK, Lindor NM, Young JP, Macrae FA, Young GP, Williamson E, et al. Risks of primary extracolonic cancers following colorectal cancer in lynch syndrome. J Natl Cancer Inst 2012;104(18):1363–1372. doi:10.1093/jnci/djs351. PubMed PMID: 22933731; PubMed Central PMCID: PMC3529597.
Conde J, Silva SN, Azevedo AP, Teixeira V, Pina JE, Rueff J, et al. Association of common variants in mismatch repair genes and breast cancer susceptibility: a multigene study. BMC Cancer 2009;9:344. doi:10.1186/1471-2407-9-344. PubMed PMID: 19781088; PubMed Central PMCID: PMC2761943.
Rennert G, Lejbkowicz F, Cohen I, Pinchev M, Rennert HS, Barnett-Griness O. MutYH mutation carriers have increased breast cancer risk. Cancer 2012;118(8):1989–1993. doi:10.1002/cncr.26506. PubMed PMID: 21952991.
Bogdanova N, Feshchenko S, Schurmann P, Waltes R, Wieland B, Hillemanns P, et al. Nijmegen breakage syndrome mutations and risk of breast cancer. Int J Cancer 2008;122(4):802–806. doi:10.1002/ijc.23168. PubMed PMID: 17957789.
Yu KD, Di GH, Yuan WT, Fan L, Wu J, Hu Z, et al. Functional polymorphisms, altered gene expression and genetic association link NRH:quinone oxidoreductase 2 to breast cancer with wild-type p53. Hum Mol Genet 2009;18(13):2502–2517. doi:10.1093/hmg/ddp171. PubMed PMID: 19351655.
Jupe ER, Badgett AA, Neas BR, Craft MA, Mitchell DS, Resta R, et al. Single nucleotide polymorphism in prohibitin 3′ untranslated region and breast-cancer susceptibility. Lancet 2001;357(9268):1588–1589. PubMed PMID: 11377649.
Kobayashi H, Ohno S, Sasaki Y, Matsuura M. Hereditary breast and ovarian cancer susceptibility genes (review). Oncol Rep 2013;30(3):1019–1029. doi:10.3892/or.2013.2541. PubMed PMID: 23779253.
Ruark E, Snape K, Humburg P, Loveday C, Bajrami I, Brough R, et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature 2013;493(7432):406–410. doi:10.1038/nature11725. PubMed PMID: 23242139; PubMed Central PMCID: PMC3759028.
Li W, Liu KJ, Song JS, Song R, Liu ZL. Association between RAD51 polymorphism and breast cancer susceptibility: a meta analysis. Int J Clin Exp Med 2015;8(2):2326–2333. PubMed PMID: 25932169; PubMed Central PMCID: PMC4402816.
Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 2010;42(5):410–414. doi:10.1038/ng.569. PubMed PMID: 20400964.
Kuschel B, Auranen A, McBride S, Novik KL, Antoniou A, Lipscombe JM, et al. Variants in DNA double-strand break repair genes and breast cancer susceptibility. Hum Mol Genet 2002;11(12):1399–1407. PubMed PMID: 12023982.
Acknowledgements
We also thank BGI-research center for providing access to sequence on the Hiseq 2500 and to analyze the post-NGS data.
Funding
This work was funded by following grants: the Science, Technology and Innovation Committee of Shenzhen Municipality (JCYJ20120614155459154), the Science, Technology and Innovation Committee of Shenzhen Municipality (CXZZ20140414170821163) and the Science, Technology and Innovation Committee of Shenzhen Municipality (JCYJ20160425103015129).
Availability of data and materials
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
Conception and design: XW, Manuscript writing and data analysis: WJ, Acquisition of pedigree information and blood: QQ, SS, Sample treatment, sequencing: KS, XW. All authors approved the MS for this publication.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by a ethics committee of Shenzhen Second people's Hospital, China. Written consent was obtained.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Jian, W., Shao, K., Qin, Q. et al. Clinical and genetic characterization of hereditary breast cancer in a Chinese population. Hered Cancer Clin Pract 15, 19 (2017). https://doi.org/10.1186/s13053-017-0079-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13053-017-0079-4



 stands for MUTYH c.892-2A > G variants.
stands for MUTYH c.892-2A > G variants.  stands for that MUTYH c.892-2A > G variant was not tested.
stands for that MUTYH c.892-2A > G variant was not tested.  stands for man.
stands for man.  stands for woman.
stands for woman.  and
and  stand for non-cancerous death.
stand for non-cancerous death.  stands for patient with breast cancer. The black arrow indicates the proband. (BC: Breast cancer)
stands for patient with breast cancer. The black arrow indicates the proband. (BC: Breast cancer)