Abstract
Purpose
There is a paucity of data on the spectrum and prevalence of pathogenic variants among women of African ancestry in the Northeast region of Brazil.
Methods
We performed BROCA panel sequencing to identify inherited loss-of-function variants in breast cancer susceptibility genes among 292 Brazilian women referred to a single institution cancer risk assessment program.
Results
The study included a convenient cohort of 173 women with invasive breast cancer (cases) and 119 women who were cancer-free at the time of ascertainment. The majority of the women self-reported as African-descended (67% for cases and 90.8% for unaffected volunteers). Thirty-seven pathogenic variants were found in 36 (20.8%) patients. While the spectrum of pathogenic variants was heterogeneous, the majority (70.3%) of the pathogenic variants were detected in high-risk genes BRCA1, BRCA2, PALB2, and TP53. Pathogenic variants were also found in the ATM, BARD1, BRIP1, FAM175A, FANCM, NBN, and SLX4 genes in 6.4% of the affected women. Four recurrent pathogenic variants were detected in 11 patients of African ancestry. Only one unaffected woman had a pathogenic variant in the RAD51C gene. Different risk assessment models examined performed well in predicting risk of carrying germline loss-of-function variants in BRCA1 and/or BRCA2 in breast cancer cases.
Conclusion
The high prevalence and heterogenous spectrum of pathogenic variants identified among self-reported African descendants in Northeast Brazil is consistent with studies in other African ancestry populations with a high burden of aggressive young onset breast cancer. It underscores the need to integrate comprehensive cancer risk assessment and genomic testing in the management of newly diagnosed Black women with breast cancer across the African Diaspora, enabling improved cancer control in admixed underserved and understudied populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the most commonly diagnosed cancer among women worldwide [1]. This non-communicable disease is the leading cause of female deaths in many countries, with widening disparities in outcomes among developed and developing countries. Geographic differences in incidence and mortality are due to many intrinsic (e.g., genetic) and extrinsic (e.g., environment, lifestyle) factors [2]. Despite notable differences, “one-size-fits-all” cancer control strategies are usually applied in screening and treatment within and across countries, which have led to widening disparities in breast cancer mortality and morbidity among different racial/ethnic groups [3].
This rising global burden of breast cancer in low- to middle-income countries demands innovative interventions to accelerate progress in cancer control and prevention. Through genomic analysis of breast cancer predisposition genes, the burden of inherited susceptibility to breast cancer in diverse populations can be better estimated, allowing clinical management and treatment recommendations to be tailored to the needs of high-risk women and their families [4]. With advances in high-throughput sequencing technologies, it is now possible to analyze numerous genomic regions simultaneously at greatly reduced cost. Many multi-gene panels such as the BROCA panel have been developed and applied successfully in large genetic testing studies in the United States and Europe [5, 6]. We previously reported the high prevalence of highly penetrant pathogenic variants in BRCA1, BRCA2, PALB2, and TP53 genes in consecutive women presenting with advanced breast cancer at tertiary hospitals in Nigeria, Cameroon, and Uganda [7, 8].
Black women across the African Diaspora have the worst outcomes from breast cancer of all ethnic/racial groups. Given the reported high prevalence of aggressive breast cancer in young Brazilian women [9], we sought to examine the burden of inherited breast cancer in a convenient sample of consecutive women with breast cancer ascertained in a cancer risk clinic in the State of Bahia in the Northeast region of Brazil. This region has a large population of African descendants as it remains segregated and is primarily inhabited by former descendants of slaves. The African ancestral proportion revealed through genomic admixture studies is the highest in this region in comparison with other regions of Brazil [10].
Methods
Study population and eligibility
Between 2008 and 2015, we recruited women with breast cancer referred by their primary care physicians to the Cancer Risk Assessment Program of the Serviço de Oncogenética of Laboratório de Imunologia e Biologia Molecular (ICS-UFBA). This public laboratory service is part of the Brazilian National Network of Hereditary Cancer. Women with breast cancer are usually referred to this service from private and public clinics and hospitals. Since this service is provided by a public entity, counseling was free of charge and more than 90% of patients were willing to participate in the research. To develop a reference control panel to improve interpretation of our findings, we recruited a cohort of cancer-free women who are not the relatives of the cases but were undergoing routine laboratory tests (for regular clinical checkups or for an evaluation of other diseases) in the same laboratory between 2014 and 2015.
All participants signed informed consents, and data regarding their epidemiological and clinical profiles were collected along with questionnaires administered by a research coordinator. The research protocol #1.383.884 was approved by the Brazilian National Committee of Ethics in Research (CONEP, Comissão Nacional de Ética em Pesquisa), the University of Chicago, and the University of Washington, where the sequencing was performed.
Next-generation sequencing and genomic analysis
The genomic DNA was extracted from peripheral blood using a commercial kit, the DNeasy® blood and tissue kit (QIAGEN, German). The quality and quantity of the genomic DNA was assessed with 2% agarose gel electrophoresis analysis and the Quant-iT™ PicoGreen™ dsDNA Assay Kit (Invitrogen, Thermo Scientific, USA). A total of 28 susceptibility genes were analyzed in the BROCA panel. Genes sequenced included established breast cancer genes of both high and moderate penetrance, and genes that have been suggested as candidate breast cancer genes, with varying levels of evidence: ATM, ATR, BAP1, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK1, CHEK2, CTNNA1, FAM175A, FANCM, GEN1, MRE11A, NBN, PALB2, PPM1D, PTEN, RAD51B, RAD51C, RAD51D, RECQL, RINT1, SLX4, STK11, TP53, and XRCC2.
Paired-end reads were mapped to the human genome reference hg19. Subsequently, single-nucleotide variants and small insertions and deletions were called as previously described in detail [5, 11], and copy number variants were detected as well [12]. Only variants that led to a loss of gene function or were experimentally demonstrated to damage gene function were included in further analyses. Interpretations of possible splice variants were based on in silico algorithms or on experimental results from our own work or that of others.
Statistical analysis
Descriptive analysis was performed using Epi Info™ software (CDC, Atlanta, GA, USA) and SPSS® (SPSS Inc. Chicago, IL, USA). Given the limited sample size of the study, we performed an exploratory analysis to estimate performance of different breast cancer risk assessment tools using online calculators: the Myriad Risk calculator [13], the PENN II Risk Model [14], and BRACAPRO [15], based on the BRCA1 and BRCA2 mutational profiles and clinical and epidemiological data.
Results
Clinical characteristics
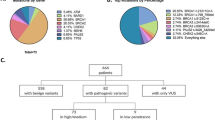
Over 90% of the study participants were from the Northeast region of Brazil, particularly from the State of Bahia. About 67.0% of the breast cancer cases self-reported as African-descended (Black), with enriched family history of cancer, including breast cancer (~ 65%) (Table 1). The mean age at breast cancer diagnosis among the cases was 44.1 ± 11.3 years while unaffected women were older, with a mean age at interview of 52.2 ± 13.6 years. The breast cancer patients were predominantly diagnosed with breast cancer only (96.5%), followed by breast and ovarian (2.3%) (Table 1). Invasive ductal carcinoma was the most common diagnosis in breast cancer patients of both African and non-African ancestry, 81.9% and 79%, respectively. The tumor subtypes were classified by immunohistochemistry for expression of hormone receptors (estrogen receptor [ER] and progesterone receptor [PR]) and human epidermal growth factor receptor 2 (HER2) (Table 2). If HER2 was classified as 2+ by immunohistochemistry, additional analysis was performed by using fluorescent in situ hybridization. The ER+/PR+/HER2- was the most common classification, followed by ER+/PR-/HER2-, triple-negative and HER2+ (41.6%, 16.2%, 15.5%, and 10.4% , respectively).
Spectrum of pathogenic variants in breast cancer susceptibility genes
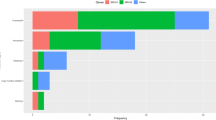
Thirty-seven loss-of-function variants (30 distinct variants) were found in 36 breast cancer patients, one of whom carried both BARD1:c.1921C>T and BRCA2:c.3860delA (Fig. 1, Table 2 and Supplementary Table 1). In the cohort of cancer-free women, only one individual carried a pathogenic variant, RAD51C:c.264_265insA. Among self-reported African-descended breast cancer patients, 24.1% (28 of 116) carried 29 pathogenic variants in ATM, BARD1, BRCA1, BRCA2, BRIP1, FAM175A, FANCM, PALB2 and TP53 genes. The majority of pathogenic variants were found in BRCA1 and BRCA2 (65.5%, 19 of 29). Four recurrent loss-of-function variants were detected in 11 African-descended breast cancer cases, BRCA1:c.3331_3334delCAAG, BRCA1:c.211A>G, BRCA2:c.1389_1390delAG and PALB2:c.1671_1674delTATT (Table 2). Pathogenic variants were found in BRCA1, BRIP1, NBN, PALB2, and SLX4 genes among eight cases of non-African ancestry (14.0%, 8 of 57), with BRCA1 and BRIP1 being the most commonly mutated genes (50%, 4 of 8) (Table 2).
Congruous with previously published annotations, we did not observe any African-specific pathogenic variants, and the majority of tumors arising in BRCA1 and BRCA2 carriers were HR- and HR+, respectively [16] (Supplementary Fig. 1). In Supplementary Fig. 2, we describe in detail the BRCA1 and BRCA2 mutational spectrum of the study population compared with other Black women across the African Diaspora and White women in the literature [17,18,19,20,21,22].
Using different risk models for pathogenic BRCA1 and BRCA2, we observed that the predicted risk was higher in breast cancer cases carrying germline loss-of-function variants in BRCA1 and/or BRCA2 than in other breast cancer genes, or those carrying wild-type genes (Supplementary Table 2). The data revealed that these risk models can discriminate high-risk from low-risk women (Supplementary Fig. 3), supporting the use of these models in this population.
Discussion
Little is known about the genetic susceptibility to breast cancer of African-descended Brazilian women, an understudied population. As there is a high degree of genetic admixture among Brazilian populations [23,24,25], we sought to study the genetic susceptibility to breast cancer in one of the largest African-descended populations in Latin America, the inhabitants of the State of Bahia in the Northeast region of Brazil.
Using the validated BROCA panel, we identified 36 distinct pathogenic variants in 30 patients with breast cancer cases and only one pathogenic variant in cancer-free women. As observed in previous studies worldwide [4, 22, 26], BRCA1 and BRCA2 were the most frequently mutated genes in breast cancer patients, and their mutation frequencies in women from the Northeast region of Brazil are closer to that in Africans than the reported frequencies in African-Americans and women of European ancestry (Table 3). We compared our results with the BRCA1 and BRCA2 mutational spectrum reported by the Brazilian Consortium of Hereditary Cancer [18], as well as other studies [17, 19,20,21,22]. Among sixteen BRCA1 and BRCA2 pathogenic variants detected in African-descended breast cancer patients in this study, twelve were documented globally, five were found in self-reported African ancestry individuals in non-African countries/regions, one was found in self-reported African ancestry individuals in African countries, eight were previously reported among Brazilians, and one was newly identified (Supplementary Fig. 2). Our observation that the BRCA1/2 pathogenic variants found in Blacks in Northern Brazil were not unique to the population agrees with the finding of a previous study by Friebel et al. [21]. BRCA1/2 pathogenic variants in Africans are allelic heterogeneous with low frequencies [7, 8, 27]; however, the Friebel et al. study showed that variants identified in Africans were also reported in non-African populations [21]. The possible explanations could be: (1) to date, breast cancer mutation surveys have been done primarily in individuals of European ancestry, which increases the probability of detecting variants also found in African populations; (2) the “out of Africa” theory of early human migrations and the diversity in the African Diaspora [27] suggest that some variants found in non-African populations could be of African origin; and (3) some African-specific variants in highly admixed populations like Brazilians might not be captured due to the limited sample size of the present study.
In this study, other genes frequently mutated were the high-risk gene PALB2, as well as ATM and BRIP1, each found in 1.7% of the cases. Although the prevalence of pathogenic variants in these genes varies in breast cancer patients of African ancestry [6, 7], it confirms that genes involved in DNA repair pathways are the major contributors to inherited breast cancer. Thus, their critical role in understudied populations with high burden of young onset breast cancer deserves further examination. In the South and Southeast regions of Brazil, TP53 is the third gene most frequently mutated among breast cancer patients. Among these patients, most carry TP53:c.1010G>A, which has a known founder effect in those regions of Brazil. However, this pathogenic variant was quite rare (0.6%, 1 of 173) among the breast cancer cases from the Northeast region of Brazil in our study (Fig. 1 and Table 2).
The spectrum of founder mutations found in our cohort reflects the degree of ancestral admixture within the State of Bahia, where there is a significant history of immigration from Spain, Central Europe, and West Africa [23, 28, 29]. Four recurrent variants were discovered among unrelated African-descended breast cancer patients: BRCA1:c.3331_3334delCAAG, BRCA1:c.211A>G, BRCA2:c.1389_1390delAG, and PALB2:c.1671_1674delTATT. The first two were previously described in Spanish descendants [30, 31] and in the Northeast Brazilian population [29]. BRCA2:c.1389_1390delAG was described in diverse populations in Central Europe [32,33,34], while PALB2:c.1671_1674delTATT remains uncharacterized and was documented only twice in ClinVar. To confirm whether this PALB2 pathogenic variant has a founder effect, further studies are needed to evaluate the carriers’ haplotypes. In addition, we observed variants that are recurrent in African populations: ATM:c.7913G>A [35, 36], BRCA1:c.815_824dupAGCCATGTGG [37, 38] and FAM175A:c.1011delA [39]. In addition, in the present study, cancer risk assessment tools like Myriad Risk, PENN II Risk, and BRCAPRO demonstrated an overall moderate efficiency at detecting high-risk individuals (Supplementary Table 2 and Supplementary Fig. 3). Larger population studies are needed to validate our findings to further improve the utility of risk prediction models in diverse populations.
This study has several limitations. First, the sample size is relatively small, which limits the study’s power to detect variants with lower frequencies. Second, the recruitment of the participants was clinic based and therefore may not represent true population frequencies. Lastly, the BROCA gene panel was not designed to include ancestry informative markers in the assay; therefore, we were unable to perform genetic ancestry analysis. An advanced targeted gene panel with ancestry informative markers included, or accompanied with a genotyping assaying capturing that information, may improve the risk assessment based on a person's ethnic background.
In summary, this study sought to investigate whether the high mutation rates observed in Nigeria, Cameroon, and Uganda are also present in Brazilian women of African ancestry using a validated multi-gene panel. As costs of genomic testing continues to drop, this study provides additional evidence in support of broader access to genetic testing for previously underserved and understudied Black women at high risk of young onset and aggressive forms of breast cancer. The fact that one in five patients carried a loss-of-function variant in BRCA1, BRCA2, or another breast cancer gene with a highly heterogeneous mutational spectrum underscores the importance of utilizing next-generation sequencing-based testing to develop screening and risk-reducing strategies in Northeast Brazil.
Data availability
All data described and analyzed here are available upon request.
Code availability
Not applicable.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424. https://doi.org/10.3322/caac.21492
Servick K (2014) Breast cancer. Breast cancer: a world of differences. Science 343(6178):1452–1453. https://doi.org/10.1126/science.343.6178.1452
Harford JB (2011) Breast-cancer early detection in low-income and middle-income countries: do what you can versus one size fits all. Lancet Oncol 12(3):306–312. https://doi.org/10.1016/S1470-2045(10)70273-4
Felix GES, Zheng Y, Olopade OI (2018) Mutations in context: implications of BRCA testing in diverse populations. Fam Cancer 17(4):471–483. https://doi.org/10.1007/s10689-017-0038-2
Walsh T, Lee MK, Casadei S et al (2010) Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci USA 107(28):12629–12633. https://doi.org/10.1073/pnas.1007983107
Churpek JE, Walsh T, Zheng Y et al (2015) Inherited predisposition to breast cancer among African American women. Breast Cancer Res Treat 149(1):31–39. https://doi.org/10.1007/s10549-014-3195-0
Zheng Y, Walsh T, Gulsuner S et al (2018) Inherited breast cancer in Nigerian women. J Clin Oncol 36(28):2820–2825. https://doi.org/10.1200/JCO.2018.78.3977
Adedokun B, Zheng Y, Ndom P et al (2019) Prevalence of inherited mutations in breast cancer predisposition genes among Uganda and Cameroon Women. Cancer Epidemiol Biomark Prev 29(2):359–367. https://doi.org/10.1158/1055-9965.EPI-19-0506
Orlandini LF, Antonio M, Espreafico CR Jr et al (2021) Epidemiological analyses reveal a high incidence of breast cancer in young women in Brazil. JCO Glob Oncol 7:81–88. https://doi.org/10.1200/GO.20.00440
Moura RR, Coelho AV, Balbino Vde Q, Crovella S, Brandao LA (2015) Meta-analysis of Brazilian genetic admixture and comparison with other Latin America countries. Am J Hum Biol 27(5):674–680. https://doi.org/10.1002/ajhb.22714
Walsh T, Casadei S, Lee MK et al (2011) Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci USA 108(44):18032–18037. https://doi.org/10.1073/pnas.1115052108
Nord AS, Lee M, King MC, Walsh T (2011) Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genomics 12:184. https://doi.org/10.1186/1471-2164-12-184
Frank TS, Deffenbaugh AM, Reid JE et al (2002) Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol 20(6):1480–1490. https://doi.org/10.1200/JCO.2002.20.6.1480
Lindor NM, Johnson KJ, Harvey H et al (2010) Predicting BRCA1 and BRCA2 gene mutation carriers: comparison of PENN II model to previous study. Fam Cancer 9(4):495–502. https://doi.org/10.1007/s10689-010-9348-3
Berry DA, Iversen ES Jr, Gudbjartsson DF et al (2002) BRCAPRO validation, sensitivity of genetic testing of BRCA1/BRCA2, and prevalence of other breast cancer susceptibility genes. J Clin Oncol 20(11):2701–2712. https://doi.org/10.1200/JCO.2002.05.121
Mavaddat N, Barrowdale D, Andrulis IL et al (2012) Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA). Cancer Epidemiol Biomarkers Prev 21(1):134–147. https://doi.org/10.1158/1055-9965.EPI-11-0775
Silva FC, Lisboa BC, Figueiredo MC et al (2014) Hereditary breast and ovarian cancer: assessment of point mutations and copy number variations in Brazilian patients. BMC Med Genet 15:55. https://doi.org/10.1186/1471-2350-15-55
Palmero EI, Carraro DM, Alemar B et al (2018) The germline mutational landscape of BRCA1 and BRCA2 in Brazil. Sci Rep 8(1):9188. https://doi.org/10.1038/s41598-018-27315-2
Rebbeck TR, Friebel TM, Friedman E et al (2018) Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum Mutat 39(5):593–620. https://doi.org/10.1002/humu.23406
Cotrim DP, Ribeiro ARG, Paixao D et al (2019) Prevalence of BRCA1 and BRCA2 pathogenic and likely pathogenic variants in non-selected ovarian carcinoma patients in Brazil. BMC Cancer 19(1):4. https://doi.org/10.1186/s12885-018-5235-3
Friebel TM, Andrulis IL, Balmana J et al (2019) BRCA1 and BRCA2 pathogenic sequence variants in women of African origin or ancestry. Hum Mutat 40(10):1781–1796. https://doi.org/10.1002/humu.23804
Palmer JR, Polley EC, Hu C et al (2020) Contribution of germline predisposition gene mutations to breast cancer risk in African American women. J Natl Cancer Inst 112(12):1213–1221. https://doi.org/10.1093/jnci/djaa040
Lima-Costa MF, Rodrigues LC, Barreto ML et al (2015) Genomic ancestry and ethnoracial self-classification based on 5,871 community-dwelling Brazilians (The Epigen Initiative). Sci Rep 5:9812. https://doi.org/10.1038/srep09812
Magalhaes da Silva T, Sandhya Rani MR, de Oliveira Costa GN et al (2015) The correlation between ancestry and color in two cities of Northeast Brazil with contrasting ethnic compositions. Eur J Hum Genet 23(7):984–989. https://doi.org/10.1038/ejhg.2014.215
Felix GES, Abe-Sandes K, Bonfim TM et al (2010) Ancestry informative markers and complete blood count parameters in Brazilian blood donors. Rev Bras Hematol Hemoter 32(4):282–285. https://doi.org/10.1590/S1516-84842010005000074
Hu C, Hart SN, Gnanaolivu R et al (2021) A population-based study of genes previously implicated in breast cancer. N Engl J Med 384(5):440–451. https://doi.org/10.1056/NEJMoa2005936
Fackenthal JD, Olopade OI (2007) Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat Rev Cancer 7(12):937–948. https://doi.org/10.1038/nrc2054
Kehdy FS, Gouveia MH, Machado M et al (2015) Origin and dynamics of admixture in Brazilians and its effect on the pattern of deleterious mutations. Proc Natl Acad Sci U S A 112(28):8696–8701. https://doi.org/10.1073/pnas.1504447112
Felix GE, Abe-Sandes C, Machado-Lopes TM et al (2014) Germline mutations in BRCA1, BRCA2, CHEK2 and TP53 in patients at high-risk for HBOC: characterizing a Northeast Brazilian Population. Hum Genome Var 1:14012. https://doi.org/10.1038/hgv.2014.12
Blay P, Santamaria I, Pitiot AS et al (2013) Mutational analysis of BRCA1 and BRCA2 in hereditary breast and ovarian cancer families from Asturias (Northern Spain). BMC Cancer 13:243. https://doi.org/10.1186/1471-2407-13-243
Vega A, Campos B, Bressac-De-Paillerets B et al (2001) The R71G BRCA1 is a founder Spanish mutation and leads to aberrant splicing of the transcript. Hum Mutat 17(6):520–521. https://doi.org/10.1002/humu.1136
Foretova L, Machackova E, Navratilova M et al (2004) BRCA1 and BRCA2 mutations in women with familial or early-onset breast/ovarian cancer in the Czech Republic. Hum Mutat 23(4):397–398. https://doi.org/10.1002/humu.9226
Goelen G, Teugels E, Bonduelle M, Neyns B, De Greve J (1999) High frequency of BRCA1/2 germline mutations in 42 Belgian families with a small number of symptomatic subjects. J Med Genet 36(4):304–308
Machackova E, Foretova L, Lukesova M et al (2008) Spectrum and characterisation of BRCA1 and BRCA2 deleterious mutations in high-risk Czech patients with breast and/or ovarian cancer. BMC Cancer 8:140. https://doi.org/10.1186/1471-2407-8-140
Coutinho G, Mitui M, Campbell C et al (2004) Five haplotypes account for fifty-five percent of ATM mutations in Brazilian patients with ataxia telangiectasia: seven new mutations. Am J Med Genet A 126A(1):33–40. https://doi.org/10.1002/ajmg.a.20570
Demuth I, Dutrannoy V, Marques W Jr et al (2011) New mutations in the ATM gene and clinical data of 25 AT patients. Neurogenetics 12(4):273–282. https://doi.org/10.1007/s10048-011-0299-0
Zhang J, Fackenthal JD, Zheng Y et al (2012) Recurrent BRCA1 and BRCA2 mutations in breast cancer patients of African ancestry. Breast Cancer Res Treat 134(2):889–894. https://doi.org/10.1007/s10549-012-2136-z
Hall MJ, Reid JE, Burbidge LA et al (2009) BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer 115(10):2222–2233. https://doi.org/10.1002/cncr.24200
Lek M, Karczewski KJ, Minikel EV et al (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536(7616):285–291. https://doi.org/10.1038/nature19057
Acknowledgements
We would like to thank all subjects who participated in this study, as all the institutions and supporting agencies that made this work possible.
Funding
This work was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Código de Financiamento – 001) (GESF), American Cancer Society (OIO, M-CK.) and the John and Editha Kapoor Charitable Foundation (OIO, M-CK), Susan G. Komen for the Cure (SAC110026 to OIO), National Cancer Institute Specialized Programs of Research Excellence (SPORE) planning grant (P20CA233307 to OIO), Secretaria de Saúde do Estado da Bahia (ILON), and Fundação de Apoio à Pesquisa e Extensão – FAPEX (RM). YZ was supported by Paul Calabresi Career Development Award for Clinical Oncology (K12 CA139160 to OIO).
Author information
Authors and Affiliations
Contributions
GESF, RSCG, OIO, ILON, KA-S, and M-CK involved in conception and design. GESF, KA-S, ILON, RM, RSCG, M-CK, and OIO took part in financial support. GESF, JC, TMML, PC, IS, TFB, BPT, RM, KA-S, ILON, and OIO participated in provision of study materials or patients. GESF, YZ, JZ, PC, JC, and TMML involved in collection and assembly of data.TW, YZ, GESF, RSCG, and EMN contributed to data analysis and interpretation. GESF, RSCG, YZ, ES, ILON, KA-S, M-CK, and OIO performed manuscript writing. All authors contributed to final approval of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
OIO is a cofounder at CancerIQ and has equity in Tempus and 54gene. RSCG acted as a consultant for AstraZeneca, GlaxoSmithKline, and Igenomix; received speaker honoraria from AstraZeneca, Bristol Myers Squibb, GlaxoSmithKline, Merck Sharp & Dohme, Novartis and Roche outside the submitted work; and has equity in Mendelics Análise Genômica. TW consults for Color Genomics. The other authors made no disclosures.
Ethical approval
This research was approved by research ethics committees or institutional review boards of all participating institutions in Brazil and the United States.
Consent to participate
All studied subjects gave informed written consent upon enrollment in the study.
Consent for publication
All studied subjects and researchers involved in this study consent to publishing the data.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10549_2022_6560_MOESM1_ESM.eps
Supplementary file1 (EPS 80 kb) Supplementary Figure 1.Breast cancer subtype distribution according to genetic mutational profile in BROCA panel. Breast cancer subtypes: HR-/HER2-, HR-/HER2+, HR+/HER2-, HR+/HER2+, HR-/HER2?, HR+/HER2?, and Missing. HER2, human epidermal growth factor receptor 2; HER2?, HER2 status unknown; HR, hormone receptor.
10549_2022_6560_MOESM2_ESM.eps
Supplementary file2 (EPS 40 kb) Supplementary Figure 2. BRCA1 and BRCA2 mutational spectrum in breast cancer cases from the Northeast region of Brazil and other populations worldwide. SRAA, self-reported African ancestry.
10549_2022_6560_MOESM3_ESM.eps
Supplementary file3 (EPS 70 kb) Supplementary Figure 3. Performance of risk prediction models. We divided our study participants into three genomically stratified groups: high-risk cases (who carried pathogenic variants in breast cancer genes), low-risk cases (who did not carry any pathogenic variants), and cancer-free women. (a) The values of AUC of the risk prediction tools improved slightly for discriminating the high-risk cases from low-risk patients. (b) When the high-risk group was compared with low-risk group and cancer-free women, the power of the risk models to discriminate high-risk individuals from low-risk was increased.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Felix, G.E.S., Guindalini, R.S.C., Zheng, Y. et al. Mutational spectrum of breast cancer susceptibility genes among women ascertained in a cancer risk clinic in Northeast Brazil. Breast Cancer Res Treat 193, 485–494 (2022). https://doi.org/10.1007/s10549-022-06560-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-022-06560-0