Abstract
Background
Interspecific hybridization is an effective strategy for germplasm innovation in sugarcane. Nobilization refers to the breeding theory of development and utilization of wild germplasm. Saccharum spontaneum is the main donor of resistance and adaptive genes in the nobilization breeding process. Chromosome transfer in sugarcane is complicated; thus, research of different inheritance patterns can provide guidance for optimal sugarcane breeding.
Results
Through chromosome counting and genomic in situ hybridization, we found that six clones with 80 chromosomes were typical S. officinarum and four other clones with more than 80 chromosomes were interspecific hybrids between S. officinarum and S. spontaneum. These data support the classical view that S. officinarum is characterized by 2n = 80. In addition, genomic in situ hybridization showed that five F1 clones were products of a 2n + n transmission and one F1 clone was the product of an n + n transmission in clear pedigree noble hybrids between S. officinarum and S. spontaneum. Interestingly, Yacheng 75–408 and Yacheng 75–409 were the sibling lines of the F1 progeny from the same parents but with different genetic transmissions.
Conclusions
This is the first clear evidence of Loethers, Crystallina, Luohanzhe, Vietnam Niuzhe, and Nanjian Guozhe were typical S. officinarum by GISH. Furthermore, for the first time, we identified the chromosome transmission of six F1 hybrids between S. officinarum and S. spontaneum. These findings may provide a theoretical basis for germplasm innovation in sugarcane breeding and guidance for further sugarcane nobilization.
Similar content being viewed by others
Background
Sugarcane, which belongs to the genus Saccharum in the family Poaceae and the tribe Andropogoneae, is related to Miscanthus, Sclerostachya, Erianthus, and Narenga, and constitutes the Saccharum complex. The genus Saccharum comprises six species, including Saccharum officinarum, Saccharum robustum, Saccharum spontaneum, Saccharum sinense, Saccharum barberi, and Saccharum edule [1]. Of these, S. spontaneum and S. robustum are considered to be wild species, as the others have been cultivated [2]. Except for S. edule, five other native species, including S. officinarum (2n = 80) and S. spontaneum (2n = 40–128), have played an important role in sugarcane breeding [1]. S. officinarum (which is referred as “noble” cane) is essential for sugarcane breeding program, as it is the main source of alleles controlling high sugar content and almost all modern sugarcane cultivars contain its lineage [3]. Typically, S. officinarum have 2n = 80 chromosomes [4], with a basic chromosome number of x = 10 [5]. S. spontaneum is a wild species characterized by high stress-resistance, and then is the most valuable wild germplasm resources in the genus Saccharum [6]. It has a wide range of chromosome numbers, ranging from 2n = 40 to 128 [7, 8]. Recently, research on S. sinense and S. barberi has shown that they are derived from natural interspecific hybridization between S. officinarum and S. spontaneum [9]. Furthermore, all modern sugarcane cultivars were hybrids between S. officinarum and S. spontaneum in the twentieth century [10]. The first artificial interspecific hybrids between these two species were created to overcome disease outbreaks and were followed by repeated backcrossing using S. officinarum as the recurrent female parent to restore high sucrose content. This procedure is referred as “nobilization”.
Interspecific hybridization is an innovative and effective method for sugarcane breeding. This strategy allows for increasing stress-resistance from S. spontaneum, as well as maintaining high sugar genes from S. officinarum, which promote the genetic improvement process [3]. Through the process of sugarcane nobilization, utilization of diverse clones of S. officinarum and S. spontaneum has been proposed as a way to introduce genetic diversity [11, 12]. While a large number of germplasm resources are available for exploitation, a limited understanding of the quantitative aspects of nobilization makes the parent selection process for nobilization difficult. In 1922, Bremer discovered the classical cytological peculiarity of 2n chromosome transmission from S. officinarum in interspecific crosses with S. spontaneum [5]. Later studies verified his work and further demonstrated that the same process occurs in BC1 when S. officinarum is used as the female parent [13]. Endoduplication, or fusion of two nuclei following the second meiosis, has been proposed by Bhat and Gill to explain this peculiar chromosome transmission [14]. However, Roach found that n + n transmission occurs in crosses between S. officinarum and S. spontaneum with 2n = 80, but seldom occurs in crosses between S. officinarum and S. spontaneum with 2n = 64 or 96 [15].
Modern sugarcane cultivars are derived from intercrossing between the first nobilized hybrids of a few parental clones with chromosome numbers ranging from 100 to 130, approximately 10% of which originating from S. spontaneum [5, 15]. The accurate number of S. spontaneum chromosomes in the different cultivars is not completely understood, as is their segregation during successive crosses. This problem impedes our understanding of the exact genetic contribution of S. spontaneum to sugarcane cultivars. To innovate germplasm in sugarcane breeding, study on chromosome composition of the progenies between S. officinarum and S. spontaneum in sufficient early generation is needed. Genomic in situ hybridization (GISH) is a highly efficient molecular cytogenetic tool that takes genomic DNA from one species as the labelled probe in hybridization experiments to chromosomal DNA in situ [16, 17]. The technique is mainly used to identify chromosome recombination, genetic relationship of interspecific hybrids, and chromosome transmission [5, 18]. To date, many researches had verified the accuracy and high-efficiency of the GISH technology in studying the chromosome composition and chromosomal translocation in a wide range of natural allopolyploids or artificial polyploidy progenies [19,20,21,22]. D’Hont et al., for the first time, demonstrated that GISH can be used to differentiate parental chromosomes in interspecific hybrids between BNS 3066 (S. officinarum) and SES 14 (S. spontaneum) [5, 18]; in addition, they identified n + n transmission of parental chromosomes in the interspecific F1 between S. officinarum and S. spontaneum. They also analyzed chromosomes of cultivar “R570” and found that approximately 10% originated from S. spontaneum and another approximately 10% were recombinant chromosomes, demonstrating that exchanges had occurred between chromosomes derived from S. officinarum and S. spontaneum. Recently, George Piperidis et al. used GISH to identify the occurrence of 2n + n transmission in crosses and the first backcrosses of S. officinarum and S. spontaneum [4]. GISH was also applied to identify parental genomes of an intergeneric hybrid between S. officinarum and a related wild species, Erianthus arundinaceus. These studies confirmed that the F1 and BC2 crosses resulted from an n + n chromosome transmission, while the BC1 cross resulted from a 2n + n transmission [23].
To date, most modern sugarcane cultivars are derived from a few clones of S. officinarum. The limited number of parents have leaded to narrow genetic background of sugarcane, various S. officinarum should be identified for germplasm innovation. Additionally, clear chromosome composition of early progeny between S. officinarum and S. spontaneum will provide enough valid germplasm for further sugarcane nobilization. The aim of the present study was to verify the authenticity of ten clones classified as S. officinarum via chromosome counting and GISH. Six clear pedigree noble F1 chromosome constitutions were analyzed using GISH. Our results will be applied to select the purest S. officinarum and valid germplasm for sugarcane breeding.
Results
Chromosome counting for identification of the authenticity of S. officinarum
We obtained chromosome preparations suitable for counting chromosomes in ten clones classified as S. officinarum (Table 2). The chromosomes were well spread with little cytoplasm background in all materials. Partial results are shown in Fig. 1, the rest results are shown in Additional file 1: Figure S1. The modal number of chromosomes for Muckche, Canablanca, 50uahapele, and Baimeizhe was 2n > 80, ranging from 86 to 114 (Table 2); however, in others six clones the chromosome modal number was 2n = 80.
GISH for identification of the authenticity of S. officinarum
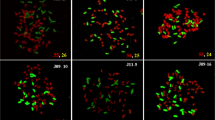
GISH was carried out on the metaphase chromosomes of ten clones classed as S. officinarum. In chromosomes, sequences homologous to S. officinarum total DNA fluoresced red and sequences homologous to S. spontaneum total DNA fluoresced green. However, due to the high homology of S. officinarum and S. spontaneum genomes, S. officinarum-derived and S. spontaneum-derived chromosomes were visualized in orange-yellow and green-yellow, respectively. The chromosomes of ten clones classed as S. officinarum were labeled in orange-yellow and green-yellow, respectively. The fluorescence of the two groups of chromosomes were differentially enhanced where their sequences were different, orange or green (Fig. 2d, e, f, and j).
GISH results of ten S. officinarum clones using biotin labelled S. officinarum genomic DNA and digoxigenin labelled S. spontaneum genomic DNA. a: Badila; b: Loethers; c: Crystallina; d: Muckche; e: Canablanca; f: 50uahapele; g: Luohanzhe; h: Vietnam Niuzhe; i: Nanjian Guozhe; j: Baimeizhe; The chromosomes of S. officinarum show orange-yellow fluorescent, while those of S. spontaneum show green-yellow fluorescent
In Badila, Loethers, Crystallina, Luohanzhe, Vietnam Niuzhe, and Nanjian Guozhe clones, all chromosomes fluoresced orange-yellow, indicating that the red signals were stronger than the green signals (Fig 2a, b, c, g, h, and i). These materials derived from only S. officinarum lineage. However, according to the color, the chromosomes of Muckche, Canablanca, 50uahapele, and Baimeizhe can be identified as two groups, orange-yellow and green-yellow (Fig 2d, e, f, and j). These orange-yellow chromosomes were derived from S. officinarum. While, the rest chromosomes fluoresced green-yellow were derived from S. spontaneum. Thus, these materials were hybrids between S. officinarum and S. spontaneum.
GISH of F1 hybrids between S. officinarum and S. spontaneum
In the six F1 hybrids analyzed, five F1 hybrids, including Yacheng 82–108, Yacheng 58–43, Yacheng 58–47, Yacheng 75–409, and Yacheng 75–419, had 2n = 112 or 120, of which 80 were derived from the S. officinarum female parent, and n = 32 or n = 40 derived from the male parents of S. spontaneum, being consistent with a typical 2n + n transmission of parental chromosomes (Table 3; Fig. 3a, b, c, d, f). However, Yacheng75–408, a sibling line of Yacheng 75–409 from the same parental combination, had 2n = 80, of which 40 were derived from the S. officinarum female parent and the other 40 were derived from the S. spontaneum male parent (Table 3; Fig. 3e). Therefore, Yacheng 75–408 is consistent with an n + n transmission of parental chromosomes.
Discussion
The authenticity of S. officinarum
Modern sugarcane cultivars have complex and unique genome structures and variable chromosome numbers. S. officinarum, which includes Badila, Black Cheribon, Crystallina, and Otaheite, has 2n = 80 chromosomes [24]; those with more than 80 chromosomes are likely to be hybrids [13, 24]. Badila is commonly used for sugarcane breeding and sugar production. Previous studies have indicated that of 31 clones in New Guinea, 29 were typical clones with chromosome number of 2n = 80 and two were atypical clones with chromosome number of 2n = 116 and 70 [25]. Piperidis et al. showed that six atypical clones (2n > 80) belong to hybrids from S. officinarum and S. spontaneum, indicating that more than 80 chromosome clones may not have originated from a pure S. officinarum [4]. In our study, the chromosome numbers of 50uahapele (2n ≈ 86), Muckche (2n ≈ 142), Baimeizhe (2n ≈ 104) and Canablanca (2n ≈ 114) were exceeded 80. Then, using GISH, we demonstrated that these cultivars were hybrids with a portion of chromosomes derived from S. spontaneum. Hence, these results were consistent with previous reports that S. officinarum may be characterized by 2n = 80 [26]. Furthermore, these differential typical S. officinarum will broaden the narrow genetic of sugarcane and provide larger pure S. officinarum for selecting cross parents in nobilization.
Chromosome transmission in F1 hybrids between S. officinarum and S. spontaneum
Interspecific hybridization had proved to be a major breakthrough for germplasm innovation in sugarcane breeding. POJ2878 is one of the most successful example in nobilization that has been widely applied [27]. However, the practical chromosome transmission is crucial for obtaining an ideal species with higher sugar, higher yield, and greater stress-resistance in nobilization. Diversity of chromosome transmission in F1 hybrids had deeply affected the efficiency of sugarcane breeding. Different genetic inheritance would lead to diverse traits of the progeny. Cytogenetic studies have demonstrated that 2n + n chromosome transmission can occur in crosses between S. officinarum (female) and S. spontaneum (male); this was also confirmed by Piperidis [4]. The 2n + n transmission is key to the nobilization process since it accelerates return to the sugar-producing type. However, the results of chromosome counting showed that n + n transmission often occurs with crosses of S. spontaneum with 2n = 80 as a male parent and seldom in crosses of S. spontaneum with 2n = 64 and 96 [15]. D’Hont et al. revealed n + n transmission of parental chromosomes by using GISH to analyze an interspecific hybrid between S. officinarum (2n = 80) and S. spontaneum (2n = 64) [5]. Here, we confirmed that four F1 clones of different series (S. spontaneum with 2n = 80 or 2n = 64 as male parents) were 2n + n. Furthermore, two different nobilization F1 clones of the same series had two different transmissions simultaneously, 2n + n or n + n, with S. spontaneum (2n = 80) as the parent. Altogether, these results concluded that diverse transmissions, 2n + n or n + n, will occurs in two different ploidy S. spontaneum (2n = 80 or 2n = 64 as male parents). Therefore, different ploidy S. spontaneum have no influence on the type of chromosome transmission (2n + n versus n + n). Furthermore, nobilization may produce different frequencies of n + n, 2n + n, and aneuploid offspring in larger numbers of F1 clones.
Many studies have shown that most F1 crosses and BC1 backcrosses result in chromosome doubling of the noble parent S. officinarum in transmission with the 2n chromosome [6,7,8, 28]. Although 2n + n is the main chromosome transmission in nobilization, there are also cases of n + n transmission [4, 5, 29]. Even more, in our study, we found that the differential transmissions in the same parents using GISH. Indeed, chromosome transmission is complex in sugarcane and further studies should be performed to guide optimized sugarcane breeding. The 2n + n chromosome transmission in interspecific crosses is considered an important factor in the rapid breakthrough that interspecific hybridization has provided to sugarcane breeding, leading to a rapid reduction in the proportion of chromosomes from wild species of hybrids and subsequent backcrosses to rapidly recover clones with highest sugar content [3, 30].
Conclusions
Using GISH, this is the first direct evidence that Loethers, Crystallina, Luohanzhe, Vietnam Niuzhe, and Nanjian Guozhe with 80 chromosomes were typical S. officinarum; while 50uahapele, Muckche, Baimeizhe and Canablanca with more than 80 chromosomes were interspecific hybrids between S. officinarum and S. spontaneum. Additionally, GISH analysis demonstrated that five F1 hybrids between S. officinarum and S. spontaneum, Yacheng 82–108, Yacheng 58–43, Yacheng 58–47, Yacheng 75–409, and Yacheng 75–419 were products of a 2n + n transmission; while, Yacheng 75–408 was the product of an n + n transmission. Although Yacheng 75–408 and Yacheng 75–409 are the sibling lines of the different F1 progeny with the same parents, there was a large difference in chromosome numbers that led to different patterns of chromosome inheritance. The results of this study support previous reports that S. officinarum may be characterized by 2n = 80 and provide more useful molecular cytogenetic information for the larger germplasm resources of S. officinarum. Futhermore, clear chromosome composition of early progenies between S. officinarum and S. spontaneum will provide guidance for further sugarcane nobilization.
Methods
Plant materials and DNA extraction
In this study, ten experimental materials classified as S. officinarum were used, including Badila, Loethers, Crystallina, 50uahapele, Muckche, Canablanca, Luohanzhe, Vietnam Niuzhe, Nanjian Guozhe, and Baimeizhe. Of these, 50uahapele, Canablanca, Baimeizhe were provided by the research Institute Ruili Station of Yunnan Agriculture Science Academy; the Sugarcane Research Institute of Yunnan Agriculture Science Academy provided the rest materials. The Hainan Sugarcane Breeding Station, Guangzhou Sugarcane Industry Research Institute provided six F1 clones between S. officinarum and S. spontaneum for nobilization (Table 1). All plant materials used in this study were grown in the germplasm resources nursery at the Fujian Agriculture and Forestry University. Leaf tissues from the above materials were ground in liquid nitrogen and stored at − 80 °C. Total genomic DNA was extracted from young leaves following CTAB methodology [31].
Chromosome preparation
Root tips were obtained from ten clones classified as S. officinarum and six clones of F1 between S. officinarum and S. spontaneum. Meristem of root-tips were treated with saturated p-dichlorobenzene solution for 1.5 h at 25 °C. The root tips were then fixed in 3:1 (v/v) ethanol: acetic acid solution for 24 h and successive eluted in ethanol solution (75, 95 and 100% ethanol), finally kept at − 20 °C with 75% ethanol solution. The fixed roots were washed in water and digested in an enzyme solution (4% Onozuka R10 cellulose, 0.5% pectolyase Y-23 and 0.5% pectinase) for 4 h at 37 °C. The digestive meristematic cells were squashed on the clear slide in 20 μL of 3:1 (v/v) ethanol: acetic acid. Slides were stored at − 20 °C.
Probe labelling
Probes were labelled using a Nick-translation kit with biotin-dUTP (Roche, Germany) and digoxigenin (Roche, Germany). For in situ hybridization, 100 ng/μL of Badila (S. officinarum) genomic DNA, labeled with biotin-dUTP, and 100 ng/μL Yunnan 75–2-11 (S. spontaneum) genomic DNA, labeled with digoxigenin were used as probes.
Genomic in situ hybridization (GISH)
GISH technique were performed as described previously by D’Hont et al. [32] with moderate improvement. The denaturing solution included 70% formamide in 2× SSC. Slides were denatured in this solution for 3 min at 80 °C. Dehydration was performed in cold ethanol and slides were then air dried at room temperature. The probe mixture including hybridization buffer (50% formamide, 2× SSC, 10% dextransulfate) and 200 ng labeled probe after denaturation for 10 min at 97 °C was applied to each slide and incubated for 20 h at 37 °C in a humid dark box. The high stringency conditions of post-hybridization washes were carried out with 2 × SSC for 8 min at 42 °C, a second wash in 50% formamide, 2 × SSC, pH 7.0, for 3 × 8 min at 42 °C, followed by a rinse in 2 × SSC for 8 min at room temperature and a final wash in 0.1 × SSC for 3 × 8 min at 55 °C. The biotin-labelled probe was detected with avidin-conjugated Texas red and the digoxigenin-labelled probe was detected with FITC (fluorescein isothiocyanate)-conjugated anti-digoxigenin antibody. Slides then were counterstained with 4′, 6-diamidino-2-phenylindole (DAPI) in a Vectashield anti-fade solution (Vector Laboratories, Burlingame, CA). GISH signals were captured using the AxioVision measurement module of AxioScope A1 Imager fluorescent microscope (Zeiss, Germany).
Chromosome counting
The metaphase chromosomes of the above materials were captured using phase contrast microscope (Fig. 1) or fluorescence microscope (Additional file 1: Figure S1; Figs. 2 and 3). The number of chromosome was counted using the program Image-pro plus 6.0 (Media Cybernetics). Results were presented as the modal number (occurred the most times among different cells in each clone) and the number of cells observed at least 30 cells for each clone (Tables 2 and 3). Additionally, at least three materials in one generation had been studied in six F1 clones.
Abbreviations
- DAPI:
-
4′, 6-diamidino-2-phenylindole
- FISH:
-
Fluorescence in situ hybridization
- FITC:
-
Fluorescein isothiocyanate
- GISH:
-
Genomic in situ hybridization
References
Irvine JE. Saccharum species as horticultural classes. Theor Appl Genet. 1999;98:186–94.
Guimarães CT, Sobral BWS. The Saccharum Complex: relation to other Andropogoneae. In: Janick J, editor. Plant breeding reviews. New York: Wiley; 2010. p. 269–88.
Roach BT. Nobilisation of sugarcane. Proc Int Soc Sugar Cane Technol. 1972;14:206–16.
Piperidis G, Piperidis N, D’Hont A. Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol Gen Genomics. 2010;284:65–73.
D’Hont A, Grivet L, Feldmann P, Rao S, Berding N, Glaszmann JC. Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp.) by molecular cytogenetics. Mol Gen Genomics. 1996;250:405–13.
Jianrong Z, Lianan T, Lihua D, Qingming Z, Huifen D, Lihe Y, Rudong A, Hongbo L, Yanfen J. Breeding potential of creation parents derived from China native Saccharum spontaneum in sugarcane. J Yunnan Agric Univ. 2011;26:12–9.
Jenkin MJ, Reader SM, Purdie KA, Miller TE. Detection of rDNA sites in sugarcane by FISH. Chromosom Res. 1995;3:444.
Panje RR, Babu CN. Studies in Saccharum spontaneum distribution and geographical association of chromosome numbers. Cytologia. 1960;25:152–72.
D'Hont A, Paulet F, Glaszmann JC. Oligoclonal interspecific origin of ‘North Indian’ and ‘Chinese’ sugarcanes. Chromosom Res. 2002;10:253–62.
Price S. Cytological studies in Saccharum and allied genera VII. Maternal chromosome transmission by S. officinarum in intra- and interspecific crosses. Int J Plant Sci. 1961;122:298–305.
Walker DIT. Utilization of noble and Saccharum spontaneum germplasm in the West Indies. Proc Int Sot Sug Cane Technol. 1971;14:224–32.
Daniels J. Improving sugarcane breeding methods to increase yields. Proc tnt Sot Sug Cane Technol. 1965;12:742–5.
Bremer G. Problems in breeding and cytology of sugar cane. Euphytica. 1961;10:59–78.
Bhat SR, Gill SS. The implication of 2n egg gametes in nobilisation and breeding of sugarcane. Euphytica. 1985;34:377–84.
Roach BT. Cytological studies in Saccharum. Chromosome transmission in interspecific and intergeneric crosses. Proc Int Soc Sugar Cane Technol. 1969;13:901–20.
Raina SN, Rani V. GISH technology in plant genome research. Methods Cell Sci. 2001;23:83–104.
Schwarzacher T, Leitch AR, Bennett MD, Heslop-Harrison JS. In situ localization of parental genomes in a wide hybrid. Ann Bot-London. 1989;64:315–24.
Piperidis N. GISH: Resolving Interspecific and Intergeneric Hybrids. In: Besse P, editor. Molecular Plant Taxonomy: Methods and Protocols. Totowa, NJ: Humana Press; 2014. p. 325–36.
An D, Zheng Q, Zhou Y, Ma P, Lv Z, Li L, Li B, Luo Q, Xu H, Xu Y. Molecular cytogenetic characterization of a new wheat-rye 4R chromosome translocation line resistant to powdery mildew. Chromosom Res. 2013;21:419–32.
Harper J, Armstead I, Thomas A, James C, Gasior D, Bisaga M, Roberts L, King I, King J. Alien introgression in the grasses Lolium perenne (perennial ryegrass) and Festuca pratensis (meadow fescue): the development of seven monosomic substitution lines and their molecular and cytological characterization. Ann Bot. 2011;107:1313–21.
Mestiri I, Chague V, Tanguy AM, Huneau C, Huteau V, Belcram H, Coriton O, Chalhoub B, Jahier J. Newly synthesized wheat allohexaploids display progenitor-dependent meiotic stability and aneuploidy but structural genomic additivity. New Phytol. 2010;186:86–101.
Zhang P, Li WL, Friebe B, Gill BS. The origin of a “zebra” chromosome in wheat suggests nonhomologous recombination as a novel mechanism for new chromosome evolution and step changes in chromosome number. Genetics. 2008;179:1169–77.
Piperidis N, Chen JW, Deng HH, Wang LP, Jackson P, Piperidis G. GISH characterization of Erianthus arundinaceus chromosomes in three generations of sugarcane intergeneric hybrids. Genome. 2010;53:331–6.
Heinz DJ. Sugarcane improvement through breeding. In: Heinz DJ, editor. Plant Breeding Reviews. Amsterdam: Elsevier; 1987. p. 15–118.
Sobhakumari VP. New determinations of somatic chromosome number in cultivated and wild species of Saccharum. Caryologia. 2013;66:268–74.
Sreenivasan TV, Ahloowalia BS, Heinz DJ. Cytogenetics. In: Heinz DJ, editor. Sugarcane improvement through breeding. New York: Elsevier; 1987. p. 211–53.
Lam E, James Shine JR, Silva JD, Lawton M, Bonos S, Calvino M, Carrer H, Silva-Filho MC, Glynn N, Helsel Z. Improving sugarcane for biofuel: engineering for an even better feedstock. GCB Bioenergy. 2009;1:251–5.
Waclawovsky AJ, Sato PM, Lembke CG, Moore PH, Souza GM, Davies M, Campbell M, Henry R. Sugarcane for bioenergy production: an assessment of yield and regulation of sucrose content. Plant Biotechnol J. 2010;8:263–76.
Price S. Cytological studies in Saccharum and allied genera. III. Chromosome numbers in interspecific hybrids. Int J Plant Sci. 1957;118:146–59.
Roach BT. Evaluation and use of sugarcane germplasm. Proc Int Soc Sugar Cane Technol. 1986;1:492–503.
Jr SC, Via LE. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. BioTechniques. 1993;14:748–50.
D’Hont A. Determination of basic chromosome numbers in the genus Saccharum by physical mapping of ribosomal RNA genes. Genome. 1998;41:221–5.
Acknowledgements
We thank the Sugarcane Research Institute of Yunnan Agriculture Science Academy, the Research Institute Ruili Station of Yunnan Agriculture Science Academy and the Hainan Sugarcane Breeding Station, Guangzhou Sugarcane Industry Research Institute for providing the plant materials used in this study. We greatly appreciate Bioscience Editing Solutions for critically reading this paper and providing helpful suggestions.
Funding
This work was funded by the Natural Science Foundation of Fujian Province of China (2016 J01094, http://yxmgl.fjkjt.gov.cn/) and supported by the earmarked fund for the Modern Agriculture Technology of China (CARS-20-1-5), the Natural Science Foundation of Guangdong Province of China (2015A030310286), and the science and technology major project of the Fujian Province of China (2015NZ0002–2, http://yxmgl.fjkjt.gov.cn/).
Availability of data and materials
The datasets supporting the conclusions of this manuscript are included within the article and its additional files.
Author information
Authors and Affiliations
Contributions
FY, PW, XL, ZD, and LX designed the study. FY, PW, and XL conducted the experiments. FY, PW, XL, YH, QW, LL, YJ, XL, ZD, JW, YY, RC, MZ, and LX analyzed the results. YH, PW, YH, ZD, and LX wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
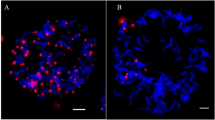
Figure S1. The metaphase chromosomes of five clones of sugarcane. A: Lothers; B: Crystallina; C: Canablanca; D: Vietnam Niuzhe; E: Nanjian Guozhe. (TIF 9259 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yu, F., Wang, P., Li, X. et al. Characterization of chromosome composition of sugarcane in nobilization by using genomic in situ hybridization. Mol Cytogenet 11, 35 (2018). https://doi.org/10.1186/s13039-018-0387-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-018-0387-z