Abstract
Background
Osteoarthritis (OA) is the most common arthritic disease in humans, affecting the majority of individuals over 65 years of age. The aim of this study is to identify the gene expression profile specific to subchondral bone in OA by comparing the different expression profiles in experimental and sham-operation groups.
Methods
Gene expression profile GSE30322 was downloaded from the Gene Expression Omnibus (GEO) database. Differentially expressed genes (DEGs) were obtained by limma package. And Database for Annotation, Visualization and Integrated Discovery (DAVID) databases were further used to identify the potential gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Furthermore, a protein–protein interaction (PPI) network was constructed and significant modules were extracted.
Results
Totally, 588 DEGs were identified including 199 upregulated DEGs and 389 downregulated DEGs screened in OA and sham-operation. GO showed that DEGs were significantly enhanced for ribosomal subunit export from nucleus and molting cycle. KEGG pathway analysis revealed that target genes were enriched in thiamine metabolism.
Conclusion
These key candidate DEGs that affect the progression of OA, and these genes might serve as potential therapeutic targets for OA.
Similar content being viewed by others
Introduction
Osteoarthritis (OA) is a degenerative disease characterized by the gradual degeneration of articular cartilage, joint stiffness, and loss of function [1]. It was reported that over 27 million adults are affected by OA in the USA [2]. OA is a complex pathophysiological process involving inflammation, subchondral bone modification, and osteophyte formation. Subchondral bone alteration present to the cartilage degeneration and thus more studies should be focused on the subchondral bone alteration.
Subchondral bone consists tripartite: subchondral bone plate, trabecular bone, and bone marrow space [3]. It has been stated that most of the OA patients accompanied by the alterations of the subchondral bone [4]. Subchondral bone could transport nutrients or cytokines to the overlying cartilage. Meanwhile, subchondral bone cells contacted with chondrocyte and thus influence cartilage metabolism. A better understanding of the early molecular mechanism changes of subchondral bone in vivo may contribute to elucidating the pathogenesis of OA. Therefore, it is crucial to explore the differentially expressed genes (DEGs) in vivo and thus we could revealed new targets for OA [5].
Microarray technology has been used to obtain information on the genetic alteration that occurs during many diseases [6, 7]. Here, we downloaded the gene expression profile GSE30322 from the Gene Expression Omnibus database (GEO), including gene expression data for subchondral bone samples from five medial meniscectomy and medial collateral ligament transection group and five sham-operated group. Based upon this research, identifying DEGs and enriching their functions and signaling pathways may help reveal potential targets of early OA.
Materials and methods
Gene expression microarray data
The gene expression profile GSE30322 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30322) was downloaded from the Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/geo/). GSE30322 was based on Agilent-014879 Whole Rat Genome Microarray 4x44K G4131F (Probe Name version) platform. GSE30322 dataset contained ten samples, including five bone 4 weeks post-surgery samples (E-group), and five sham-operated group (S-group) 4 weeks post-surgery samples.
DEGs in E-group and intact S-group samples
The raw data files were downloaded and then python scripts for matrix transformation were used. The analysis was carried out using Limma package from Bioconductor project. In this study, genes with P < .05 and [log fold change (FC)] > 2 were defined as DEGs. The DEGs data were then processed by R software (pheatmap package) to draw a heatmap and volcano plot.
GO and KEGG analysis of DEGs
Target genes list were submitted to the DAVID 6.8 (https://david.ncifcrf.gov/) to analyze candidate DEG functions and Kyoto Encyclopedia of Genes and Genomes (KEGG) of the overlapping genes. DEG functions, also named as Gene ontology (GO), mainly including biological process (BP), molecular function (MF), and cellular component (CC). P value less than 0.05 was considered as cut-off criterion [8,9,10].
Protein-protein interaction (PPI)
We used the online database STRING (Search Tool for the Retrieval of Interacting Genes, https://string-db.org/) to better illustrate the potential interactive relationships among the DEGs [11]. Then, the Cytoscape software was utilized for analyzing the interactions with a combined score > 0.4 (http://www.cytoscape.org/). Finally, the plug-in Molecular Complex Detection (MCODE) was used to filter the significant modules from the PPI network for the selection of hub genes (degree cut-off = 2, node score cut-off = 0.2, k-core = 2, and max. depth = 100) [12].
Results
Identification of DEGs
After analyzing, differentially expression gene profiles were obtained. Totally, 588 DEGs were identified including 199 upregulated DEGs and 389 downregulated DEGs screened in OA and sham-operation. Top 10 up-DEGs and down-DEGs were listed in Table 1 and Table 2, respectively. A box plot of the sample data is provided in Fig. 1. Volcano plot of the different genes can be obtained in Fig. 2. Moreover, we provided heatmap of the top 50 different genes between E-group and S-group (Fig. 3).
GO term enrichment analysis of DEGs
Gene Ontology (GO) showed that up-DEGs were significantly enhanced for ribosomal subunit export from nucleus, ribosome localization, regulation of hemopoiesis, negative regulation of hemopoiesis, and rRNA-containing ribonucleoprotein complex export from nucleus. Downregulated DEGs were enriched for the molting cycle, hair cycle, molting cycle process, hair cycle process, and the skin epidermis development (Table 3 and Fig. 4).
KEGG pathway analysis of DEGs
The result of KEGG pathway analysis revealed that target genes were enriched in thiamine metabolism, regulation of lipolysis in adipocytes, central carbon metabolism in cancer, estrogen signaling pathway, collecting duct acid secretion, Rap1 signaling pathway, measles, sphingolipid metabolism, drug metabolism-other enzymes, and circadian rhythm. These key pathways were showed in Table 4 and Fig. 5.
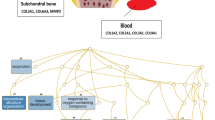
Interaction network of DEGs and core genes in the interaction network
Using data from the Cytoscape and STRING databases, the 10 hub nodes with the greatest degree of network connection were determined. The top 10 hub genes identified were RASL2-9, PSMD6, CPOX, FTH1, PYGL, GNAI1, PTPN1, RIC8A, RAB7A, LOC680441, USP4, and HIST2H2BE (Fig. 6 and Table 3). We listed top three MCODE results in Fig. 7.
Discussion
Subchondral bone remodeling is regulated by the bone resorption and bone formation, which mainly regulated by osteoclast and osteoblast respectively. Despite advancements in the understanding of the mechanism of OA, an effective method for accelerating the process remains to be identified. In this analysis, we found that 588 DEGs between E-group and S-group.
Bone remodeling is regulated by the balanced processes of osteoclast-mediated bone resorption and osteoblast-mediated bone formation [13, 14]. Disequilibrium of this balance leads to dysregulated bone tissue remodeling and can result in excessive bone loss or extra bone formation and consequent skeletal disease [15]. We identified DEGs that play roles in osteoclast and osteoblast differentiation and function in the early stages of OA in this model. Previous studies identified that an altered phenotype of subchondral osteoblasts and osteoclasts contribute to OA progress. Lamas et al. [16] reported that COL10A1 downregulation seems to have a role in the establishment of a defective and/or unstable subchondral cartilage matrix in OA disease. Ren et al. [17] used gene expression profile GSE103416 to identify the different expression genes and potential pathways. Results show that Gna13/cGMP-PKG signaling pathway was identified as a potential research target for therapy and for further understanding the development of OA.
We further performed Kegg pathways to identify the potential pathways that involving in the progress of OA. We found that Rap1 signaling pathway was the most obvious different signaling pathways. For Rap1 signaling pathway, we found nine DEGs (GNAI3/RAPGEF5/CSF1/PIK3CD/MK1/LPAR2/P2RY1/GNAS/PRKD1/RASSF5). Another potential pathway was estrogen signaling pathway. Ren et al. [17] used gene expression profile GSE103416 and found that Gna13/cGMP-PKG signaling pathway was identified as a potential research target for therapy and for further understanding the development of OA. Feng et al. [18] revealed that PDGFRB, IFNG, EGR1, FASLG, and H3F3B may be the potential targets for OA diagnosis and treatment. Liang Y al [19]. found that estrogen deficiency is closely related to the development of menopausal arthritis including OA. Estrogen acts via ER and miR-140 to inhibit the catabolic activity of proteases within the chondrocyte extracellular matrix.
We also found that Rap1 signaling pathway participated into the pathological process of OA. Zhang et al. [20] performed a gene expression analyses of subchondral bone in early experimental osteoarthritis by microarray. Results found that Alp, Igf1, Tgf β1, Postn, Mmp3, Tnfsf11, Acp5, Bmp5, Aspn, and Ihh genes that involved in the pathological process of OA, and they also performed PCR to identify these DEGs in the OA and normal cartilage. Kovács et al. [21] also revealed that the Wnt and the OPG-RANKL-RANK signaling systems, as key mediators, interact in subchondral bone remodeling in OA development, which indicated that subchondral bone remodeling also affects OA. Zhou et al. [22] revealed that matrix metalloproteinase (MMP)1, MMP3, MMP13, and prostaglandin-endoperoxide synthase 2 (PTGS2) are associated with human developmental chondrogenesis.
Conclusion
In summary, totally, 588 DEGs were identified including 199 upregulated DEGs and 389 downregulated DEGs screened in OA and sham-operation. We identified 12 core genes, including RASL2-9, PSMD6, CPOX, FTH1, PYGL, GNAI1, PTPN1, RIC8A, RAB7A, LOC680441, USP4, and HIST2H2BE. Additional experimental studies will be needed to validate our findings.
Availability of data and materials
The data was freely downloaded from the public GEO database.
Abbreviations
- OA:
-
Osteoarthritis
- DAVID:
-
Database for Annotation, Visualization and Integrated Discovery
- DEGs:
-
Differentially expressed genes
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- PPI:
-
Protein–protein interaction
- GEO:
-
Gene Expression Omnibus
- FC:
-
Fold change
- BP:
-
Biological process
- MF:
-
Molecular function
- CC:
-
Cellular component
- STRING:
-
Search Tool for the Retrieval of Interacting Genes
- MCODE:
-
Molecular Complex Detection
References
Lories RJ, Luyten FP. The bone-cartilage unit in osteoarthritis. Nat Rev Rheumatol. 2011;7:43–9. https://doi.org/10.1038/nrrheum.2010.197.
Moskowitz RW. The burden of osteoarthritis: clinical and quality-of-life issues. Am J Manag Care. 2009;15:S223–9.
Madry H, van Dijk CN, Mueller-Gerbl M. The basic science of the subchondral bone. Knee Surg Sports Traumatol Arthrosc. 2010;18:419–33. https://doi.org/10.1007/s00167-010-1054-z.
Bobinac D, Marinovic M, Bazdulj E, et al. Microstructural alterations of femoral head articular cartilage and subchondral bone in osteoarthritis and osteoporosis. Osteoarthr Cartil. 2013;21:1724–30. https://doi.org/10.1016/j.joca.2013.06.030.
Razmara E, Bitaraf A, Yousefi H, et al. Non-Coding RNAs in Cartilage Development: An Updated Review. Int J Mol Sci. 2019;20. https://doi.org/10.3390/ijms20184475.
Elgaaen BV, Olstad OK, Sandvik L, et al. ZNF385B and VEGFA are strongly differentially expressed in serous ovarian carcinomas and correlate with survival. PLoS ONE. 2012;7:e46317. https://doi.org/10.1371/journal.pone.0046317.
Al-Rekabi Z, Wheeler MM, Leonard A, et al. Activation of the IGF1 pathway mediates changes in cellular contractility and motility in single-suture craniosynostosis. J Cell Sci. 2016;129:483–91. https://doi.org/10.1242/jcs.175976.
Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–6. https://doi.org/10.1093/nar/gkj021.
Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. https://doi.org/10.1038/75556.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. https://doi.org/10.1093/nar/28.1.27.
Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–52. https://doi.org/10.1093/nar/gku1003.
Bandettini WP, Kellman P, Mancini C, et al. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J Cardiovasc Magn Reson. 2012;14:83. https://doi.org/10.1186/1532-429x-14-83.
Li G, Yin J, Gao J, et al. Subchondral bone in osteoarthritis: insight into risk factors and microstructural changes. Arthritis Res Ther. 2013;15:223. https://doi.org/10.1186/ar4405.
Aizah N, Chong PP, Kamarul T. Early Alterations of Subchondral Bone in the Rat Anterior Cruciate Ligament Transection Model of Osteoarthritis. Cartilage. 2019;1947603519878479. https://doi.org/10.1177/1947603519878479.
Martig S, Hitchens PL, Lee PVS, et al. The relationship between microstructure, stiffness and compressive fatigue life of equine subchondral bone. J Mech Behav Biomed Mater. 2019;101:103439. https://doi.org/10.1016/j.jmbbm.2019.103439.
Lamas JR, Rodriguez-Rodriguez L, Vigo AG, et al. Large-scale gene expression in bone marrow mesenchymal stem cells: a putative role for COL10A1 in osteoarthritis. Ann Rheum Dis. 2010;69:1880–5. https://doi.org/10.1136/ard.2009.122564.
Ren YM, Zhao X. Exploring the key genes and pathways of osteoarthritis in knee cartilage in a rat model using gene expression profiling. Yonsei Med J. 2018;59:760–8. https://doi.org/10.3349/ymj.2018.59.6.760.
Feng Z, Lian KJ. Identification of genes and pathways associated with osteoarthritis by bioinformatics analyses. Eur Rev Med Pharmacol Sci. 2015;19:736–44.
Liang Y, Duan L, Xiong J, et al. E2 regulates MMP-13 via targeting miR-140 in IL-1beta-induced extracellular matrix degradation in human chondrocytes. Arthritis Res Ther. 2016;18:105. https://doi.org/10.1186/s13075-016-0997-y.
Zhang R, Fang H, Chen Y, et al. Gene expression analyses of subchondral bone in early experimental osteoarthritis by microarray. PLoS ONE. 2012;7:e32356. https://doi.org/10.1371/journal.pone.0032356.
Kovacs B, Vajda E, Nagy EE. Regulatory effects and interactions of the Wnt and OPG-RANKL-RANK signaling at the bone-cartilage interface in osteoarthritis. Int J Mol Sci. 2019;20. https://doi.org/10.3390/ijms20184653.
Zhou J, Li C, Yu A, et al. Bioinformatics analysis of differentially expressed genes involved in human developmental chondrogenesis. Medicine (Baltimore). 2019;98:e16240. https://doi.org/10.1097/md.0000000000016240.
Acknowledgements
None.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
ZW designed the study and developed the retrieve strategy. HWB and JY searched and screened the summaries and titles. ZW, HWB, and JY drafted the article. All authors read and approved the final draft.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable. This paper does not involve research on humans.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, Z., Ji, Y. & Bao, Hw. Bioinformatics analysis of differentially expressed genes in subchondral bone in early experimental osteoarthritis using microarray data. J Orthop Surg Res 15, 310 (2020). https://doi.org/10.1186/s13018-020-01839-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13018-020-01839-8