Abstract
Background
Agrobacterium rhizogenes-mediated hairy root transformation provides a powerful tool for investigating the functions of plant genes involved in rhizobia-legume symbiosis. However, in the traditional identification methods of transgenic hairy roots based on reporter genes, an expensive chemical substrate or equipment is required.
Results
Here, we report a novel, low cost, and robust reporter for convenient, non-destructive, and directly visual selection of transgenic hairy roots by naked eye, which can be used in the study of rhizobia-legume symbiosis. The reporter gene AtMyb75 in Arabidopsis, encoding an R2R3 type MYB transcription factor, was ectopically expressed in hairy roots-mediated by A. rhizogenes, which induced purple/red colored anthocyanin accumulation in crop species like soybean (Glycine max (L.) Merr.) and two model legume species, Lotus japonicas and Medicago truncatula. Transgenic hairy roots of legumes containing anthocyanin can establish effective symbiosis with rhizobia. We also demonstrated the reliability of AtMyb75 as a reporter gene by CRISPR/Cas9-targeted mutagenesis of the soybean resistance to nodulation Rfg1 gene in the soybean PI377578 (Nod-) inoculated with Sinorhizobium fredii USDA193. Without exception, mature nitrogen-fixation nodules, were formed on purple transgenic hairy roots containing anthocyanin.
Conclusions
Anthocyanin is a reliable, user-friendly, convenient, non-destructive, low cost, directly visual reporter for studying symbiotic nitrogen-fixing nodule development and could be widely applied in broad leguminous plants.
Similar content being viewed by others
Background
The leguminous plants, including crop species soybean (Glycine max (L.) Merr.) and model legume species Lotus japonicus and Medicago truncatula, can establish a symbiosis relationship with rhizobia and form root nodules [1]. Rhizobia reduce atmospheric dinitrogen in root nodules to ammonia that is utilized by the host legumes, resulting in improved plant growth for sustainable agriculture [2]. Agrobacterium rhizogenes-mediated hairy root transformation for generating composite plants composed of transgenic roots and wild-type shoot provides a powerful tool for investigating the functions of plant genes involved in legume-rhizobia symbiosis [3]. However, not all the hairy roots induced by A. rhizogenes are transgenic [3, 4]. To facilitate the identification of the transgenic roots, a reporter gene on the binary vector transformed often was employed. In the traditional screening methods of transgenic roots based on reporter genes, the β-glucuronidase (GUS) activity or fluorescent proteins (such as GFP, YFP, RFP, CFP, etc.) were the most widely utilized [1, 5,6,7,8,9,10,11,12]. GUS staining assay is destructive to plant tissues, and an expensive chemical substrate (X-Gluc) is required. Furthermore, the staining buffer includes potassium ferricyanide and potassium ferrocyanide, which are detrimental to human health [4, 13]. A prominent feature of fluorescent proteins is a non-destructive and visual reporter without a requirement of an additional substrate. However, the observation of fluorescent proteins is dependent on fluorescent microscope, and automated fluorescence background of plant tissue often interferes with screening of the transgenic roots [10]. Furthermore, eyes are subject to being sore and uncomfortable to observe fluorescence. Recently, Mitiouchkina et al. [14] reported luminescence Nicotiana benthamiana plants engineered by converting ceffeic acid into luciferin that is visible to the naked eye. However, it is not a convenient operation to integrate four Neonothopanus nambi bioluminescence genes: nnluz (luciferase), nnhisps (hispidin synthase), nnh3h (hispidin-3-hydroxylase) and nncph (caffeoyl pyruvate hydrolase) into plants.
Anthocyanin belongs to flavonoids that benefit for human health and is attributable to coloration of plant organs or tissues. The Arabidopsis AtMyb75/PRODUCTION OF ANTHOCYANIN PIGMENTS 1 (PAP1, GenBank No. AY519563) encodes an R2R3 type MYB transcription factor, which regulates the production of anthocyanin [15]. To overcome the drawbacks of traditional methods for selection transgenic hairy roots, here, we developed a novel reporter gene AtMyb75/PAP1 that can be used in the study of rhizobia-legume symbiosis without interfering with nitrogen-fixing nodule development. AtMyb75/PAP1 was ectopically expressed in hairy roots-mediated by A. rhizogenes, that induced the purple/red colored anthocyanin accumulation in the legumes soybean (Glycine max (L.) Merr.) with purple hypocotyl, L. japonicus, L. corniculatus, and M. truncatula. Transgenic hairy roots of legumes containing anthocyanin can establish effective symbiosis with rhizobia. The AtMyb75/PAP1 can be served as a reliable reporter gene was further validated by targeted editing the soybean resistance to nodulation Rfg1 gene by CRISPR/Cas9 system in soybean PI377578 (Nod-) inoculated with Sinorhizobium fredii USDA193. We anticipate anthocyanin will be widely used to screen the transgenic hairy roots by A. rhizogenesis-mediated transformation in the study of rhizobia-legume symbiosis.
Methods
Plant materials and growth conditions
Soybean (Glycine max (L.) Merr.) PI 377578, and other different genotypes of soybean (listed in Additional file 1: Table S1) seeds used in this study were provided by the National Key Facility for Crop Gene Resources and Genetic Improvement (NFCIR), Institute of Crop Science, Chinese Academy of Agricultural Sciences. Lotus japonicus (Gifu-129), Lotus corniculaus L (Linn. Li’ao), and Medicago truncatula (accession R108) seeds were kept in our lab. Plants were cultivated in a growth chamber (24–26 °C, 16 h/8 h light/dark cycle).
Cloning procedure of AtMyb75/PAP1 and plasmids construction
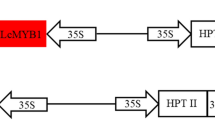
To produce an AtMyb75-overexpression vector, termed as p35SAt75 (Fig. 1a), AtMyb75 was amplified by PCR using wild-type Arabidopsis thaliana (Col-0) genomic DNA as a template by oligos At75SF (5′-GTATCGACTTTGTTCCATGGAGGGTTCG-3′) and At75SR (5′-ACGGTCGACCACAAACGCAAACA AATGTTCG-3′) (SalI restriction site underlined). The fragment AtMYB75 was digested with SalI and then ligated into the XhoI restriction enzyme site of pYGUS1305 binary vector [4] containing a GUSPlus gene driven by YAO promoter [4, 16], which can be used for identification of transgenic hairy roots. In the recombinant binary vector p35SAt75, AtMYB75 reporter gene driven by CaMV 35S promoter replaced the Hpt II (Hygromycin Phosphotransferase II) based on the backbone of pYGUS1305 [4]. To generate CRISPR/Cas9-mediated Rfg1 gene knockout vector, designated as pPG35Cas9 (Fig. 1b), the DNA fragment covering CaMV 35S promoter, AtMYB75 and left border of T-DNA was amplified with PCR primers At75E3 (5′-GCCAATTGATTGACAACGTTGCGTATTGGCTAGAGCAG-3′) and At75S1 (5′-AGCCGATTTTGAAACCGCGATGATCACAGGCAGCAACGCT-3′) using p35SAt75 as a template. The generated DNA fragment was then inserted in place of the Hpt II (Hygromycin Phosphotransferase II) reporter gene in the binary vector pHSE401 [17] which was linearized by EcoRI and SacII prior to homologous recombination. One targeted site of Rfg1 was constructed as described previously [11]. All vectors constructed were verified by sequencing. The generated binary recombinant vectors p35AtM75 and pPG35Cas9 were transformed into A. rhizogenes K599 (for soybean transformation) and ARqual strains (for L. japonicus, L. corniculatus and M. truncatula transformation) by electroporation, respectively.
A. rhizogenesis-mediated hairy root transformation and nodulation assay
Composite soybean plants were generated using one-step A. rhizogenes-mediated transformation [3]. Composite L. japonicus, L. corniculatus [18] and M. truncatula [19] plants were generated based on previously published protocols. Nodulation assay was performed as described by Fan et al. [3] in soybean, and Okamoto et al. [18] in L. japonicus. Compatible rhizobia USDA110 and microsymbiont Mesorhizobium loti MAFF303099 were used to inoculate with soybean and L. japonicus to induce nodulation, respectively [20, 21]. Nodulation phenotype was evaluated at 4 weeks post-inoculation. To water the composite plants, the sterile N-free nutrient solution containing 1 M CaCl2, 0.5 M KH2PO4, 10 mM Fe-citrate, 0.25 M MgSO4, 0.25 M K2SO4, 1 mM MnSO4, 2 mM H3BO4, 0.5 mM ZnSO4, 2 mM CuSO4, 0.1 mM CoSO4, 0.1 mM Na2MoO4) was used [22].
GUS staining assay
Histochemical staining of GUS activity qualitative assay followed the protocol [13] with some modifications. The samples were incubated in a GUS staining solution (100 mM sodium phosphate at pH 7.0, 0.1% Triton X-100, 1 mg/mL X-Gluc, 1 mM potassium ferricyanide, and 1 mM potassium ferrocyanide) in the dark at 37 °C for 2-10 h. The samples stained were rinsed in a 70% ethanol for 10-20 min.
Semi-quantitative RT-PCR
To determine whether the transformed hairy roots with anthocyanin accumulation are overexpression-AtMYB75 or not, we carried out RT-PCR analysis. The transcripts of AtMYB75 in the transformed soybean hairy roots were detected. To amplify AtMYB75, the gene-specific forward primer RTMyb75F1 (5′-TTTGTTCCATGGAGGGTTCG-3′) and reverse primer RTMyb75R1 (5′-ACCTATTCCCTAGAAGCCTATG-3′) were used and the amplification fragment covers a partial 5′ untranslated region (10-bp), a partial first exon (121-bp), the first intron (540-bp) and a partial second exon (130-bp) region. Different sizes of fragment were amplified from PCR and RT-PCR products (801-bp and 261-bp, respectively) to exclude the contamination possibility from genome DNA of the plant sample and binary vector p35AtM75 from the A. rhizogenesis. The isolation of total RNA, purification of poly (A) ± mRNA from total RNA, synthesis of first-strand cDNA were performed as previously described [23]. RT-PCR reaction was conducted according to Fan et al. [3]. The soybean GmActin was amplified and employed as an internal control using a forward primer GmActinF (5′-GAGCTATGAATTGCCTGATGG-3′) and a reverse primer GmActinR (5′-CGTTTCATGAATTCCAGTAGC-3′) [3].
Statistics
Data were analyzed using Microsoft office Excel 2016 and Data Processing System (DPS) statistical software. The averages ± standard deviations of three independent experiments were calculated. Each experiment included at least 20 composite plants. Dry weight of hairy roots was measured 4 weeks post-inoculation and dried at 105 °C for 20 min, and then 80 °C for 12 h.
Results
Directly visual selection of transgenic hairy roots-mediated by A. rhizogenes in legumes
To analyze whether ectopic expression of AtMyb75 in the roots of legumes can induce the anthocyanin accumulation, a CaMV 35S promoter-driven AtMyb75 overexpression construct, p35AtM75, was transformed into the legumes soybean (Glycine max (L.) Merr.) PI 377578, L. japonicus, L. corniculatus, and M. truncatula (accession R108) by A. rhizogenes-mediated transformation. The ectopic expression of AtMyb75 in the hairy roots induced purple/red colored anthocyanin accumulation in the transformed roots in soybean (Fig. 2a), L. corniculatus (Fig. 2b), L. japonicus (Fig. 2c, d), and M. truncatula (Fig. 2e). In contrast, anthocyanin was absent normally in these non-transformed roots (Fig. 2a–e). The purple/red transgenic roots with anthocyanin accumulation were directly visual to naked eyes without any exogenous substrate supplemented or equipment used, and readily distinguished from the white non-transgenic roots (Fig. 2a–e). The transgenic positive soybean roots were confirmed by GUS staining assay (Fig. 2f). Results showed that the purple/red-positive roots are consistent with the GUS-positive ones (Fig. 2a, f). Ectopic expression of AtMyb75 in hairy roots containing anthocyanin was further confirmed by semi-quantitative RT-PCR analysis (Additional file 2: Fig. S1).
Anthocyanin accumulation in the transgenic hairy roots of leguminous plants containing 35S:: AtMyb75. Purple/red coloration is direct visualization in the overexpression-AtMyb75 hairy roots of soybean PI377578 (a), L. corniculaus (b), L. japonicus (c, d) and M. truncatula R108 (e), respectively. Section f is the GUS stained hairy roots in section a, respectively. Transgene-positive roots are indicated by arrows. Bars = 1 cm
Evaluation the reliability of anthocyanin as a reporter in the study of rhizobia-legume symbiosis
To evaluate whether the produced anthocyanin in transgenic roots interferes with legume nodulation and can be served as a reporter applied for nitrogen-fixing nodule development, compatible rhizobia USDA110 and M. loti MAFF303099 were used to inoculate with soybean and L. japonicus to induce nodulation, respectively. Mature nitrogen-fixing nodules were formed on purple/red transgenic roots produced anthocyanin in soybean (Fig. 3a, b) and L. japonicus (Fig. 3c, d). Purple/red anthocyanin was visual to naked eyes in both transgenic roots and nodules (Fig. 3). The result indicated that transgenic roots of legumes with anthocyanin accumulation nodulated normally and could establish effective symbiosis with rhizobia. The nodule numbers per root dry weight of transgenic roots were not significantly different (p ˂ 0.05) from that of non-transgenic roots, even though the transgenic roots with a higher level of anthocyanin accumulation compared with non-transgenic roots (infected with K599 harboring pCAMBIA1305.1) were used for statistical analysis (Fig. 4). These results indicated that anthocyanin do not affect the interaction of the plant roots with rhizobia in soybean and L. japonicus.
Assessment of the nodulation ability in the roots of soybean and L. japonicas containing anthocyanin. Mature nitrogen-fixation nodules formed on purple/red transgenic roots in soybean (a, b) and L. japonicus (c, d); Pictures b and c are close-up of sections a and d marked in the white box, respectively. Composite soybean plant was generated according to the protocol [22] in Fig. 4a. Bars = 1 cm
Specific rhizobia can establish effective symbiosis with specific legumes species (or genotypes of legumes) and therefore form nitrogen-fixation nodules [11]. For instance, soybean PI377578 prevents Sinorhizobium fredii USDA193 from nodulation (Nod-) and the dominant Rfg1 in the soybean is responsible for nodulation restriction (our unpublished data). To further test the reliability of AtMyb75 as a reporter gene, Rfg1 was knockout by the CRISPR/Cas9 system in PI377578 inoculated with USDA193. Transgenic roots can form nodules only containing homozygous or bi-allelic Rfg1 mutation [11]. Mature nitrogen-fixation nodules, as expected, were successfully formed on purple/red transgenic roots produced anthocyanin (Fig. 5a, b). No nodules were formed on non-transgenic roots (Fig. 5a). The transgenic nodules were confirmed by PCR analysis and sequencing (Fig. 5c).
CRISPR/Cas9-mediated knockout of Rfg1 in the soybean PI377578 (Nod-) background inoculated with USDA193. a Mature nodules formed on overexpression-AtMyb75 hairy roots. Close-up of section a marked in the white box was shown in section b. c A sequencing identification from CRISPR/Cas9-mediated knockout of Rfg1 in the soybean PI 377578 (Nod-) background. An example of sequencing analysis of the DNA from wild type nodule (WT) and targeted site and PAM site (AGG) are shown within the box. Transgenic nodules revealed that two mutant alleles with a 52-bp (mutant 1#) and 152-bp (mutant 2#) deletion were caused, respectively. The deleted site is shown by a black arrow. Bars = 1 cm
Application of the anthocyanin as a reporter to different genotypes of soybean
To assess whether overexpression-AtMyb75 in hairy roots can induce anthocyanin accumulation in various soybean cultivars, we analyzed 86 genotypes of soybean. Visual anthocyanin was accumulated in 39 out of 86 surveyed soybean varieties. Overexpression-AtMyb75 transgenic roots in soybean cultivars with purple hypocotyl could be accumulated purple/red anthocyanin. Compared with this, there is no purple/red anthocyanin accumulation in overexpression-AtMyb75 transgenic roots induced from the soybeans with green hypocotyl (Additional file 1: Table S1). The anthocyanin accumulation in overexpression-AtMyb75 roots might be linked with purple hypocotyl in soybean. This requires further experimental testing of more different genotypes of soybean with purple and green hypocotyls.
Discussion
In this study, whether anthocyanin could be taken as a novel reporter in studying legume-rhizobia symbiosis was analyzed. Ectopic expression of AtMyb75 in soybean with purple hypocotyl, leguminous plant L. japonicas, L. corniculaus and M. truncatula resulted in purple/red anthocyanin production in transgenic hairy roots-mediated by A. rhizogenes transformation. Transgenic hairy roots were easily to be distinguished from non-transgenic roots under white light by the color conferred by the anthocyanin. The transgenic soybean and L. japonicus hairy roots containing 35S:: AtMyb75 which was accumulated with anthocyanin can form mature nodules inoculated with compatible rhizobia. The targeted mutation of Rfg1 in soybean PI377578 by CRISPR–Cas9 system further validated that anthocyanin served as a reporter is reliable. Taking these findings together, we conclude that anthocyanin can be served as a novel reporter applied in directly visual selection transgenic hairy roots-mediated by A. rhizogenes transformation in the study of rhizobia-legume symbiosis.
Anthocyanin can be synthesized in the early hairy roots with ~ 1 cm length (Fig. 2b, c, e) and persisted through later stage of nodule development (Fig. 3). These results indicated that anthocyanin provides an early-stage selection tool for the transgenic hairy roots. During preparing this paper, another study also demonstrated that the application of anthocyanin as a reporter in M. truncatula for studying nitrogen-fixing nodule development [24], which corroborates the reliability of our method. Based on our results and previous study, we conclude that anthocyanin is a user-friendly, convenient, non-destructive, low cost, directly visual ideal reporter in research on rhizobia-legume symbiosis.
In this study, we also observed that some genotypes of soybean with green hypocotyl containing overexpression-AtMyb75 can not induce the production of anthocyanin in hairy roots. This might result from no downstream or interaction genes regulated by AtMyb75 involving in synthesis of anthocyanin in the roots of these genotypes. However, anthocyanin can be synthesized by transformed with overexpression-AtMyb75 in the soybeans with purple hypocotyl tested. Anyway, till this study, anthocyanin served as a reporter can be applied in some leguminous plant species. In the future, with the elucidating of the synthesis mechanism of anthocyanin, if possible, anthocyanin can be accumulated by regulating the expression of other gene(s) involving in the synthesis of anthocyanin in those soybeans with green hypocotyl and therefore widely applied in all genotypes of soybean as a reporter.
Conclusions
Anthocyanin is a user-friendly, healthy to human and environment, convenient, non-destructive, low cost, directly visual reporter gene and suitable for transgenic selection of hairy roots-mediated by A. rhizogenes transformation in studying the nitrogen-fixation nodules development.
Availability of data and materials
All data supporting the conclusions of this article are included in this article.
Abbreviations
- CRISPR/Cas9:
-
clustered regularly interspaced short palindromic repeats-associated Cas9
- GFP/RFP/YFP/CFP:
-
Green/Red/Yellow/Cyan fluorescent protein
- GUS:
-
β-glucuronidase
- RT-PCR:
-
reverse transcriptase polymerase chain reaction
- bp:
-
base pairs
- K599-p35AtM75:
-
K599 harboring p35AtM75 vector
References
Roy S, Liu W, Nandety RS, Crook A, Mysore KS, Pislariu CI, et al. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell. 2019. https://doi.org/10.1105/tpc.19.00279.
Shine MB, Gao Q, Chowda-Reddy RV, Singh AK, Kachroo P, Kachroo A. Glycerol-3-phosphate mediates rhizobia-induced systemic signaling in soybean. Nat Commun. 2019;10:5303. https://doi.org/10.1038/s41467-019-13318-8.
Fan Y, Zhang X, Zhong L, Wang X, Jin L, Lyu S. One-step generation of composite soybean plants with transgenic roots by Agrobacterium rhizogenes-mediated transformation. BMC Plant Biol. 2020;20:208.
Fan Y, Xu F, Zhou H, Liu X, Yang X, Weng K, et al. A fast, simple, high efficient and one-step generation of composite cucumber plants with transgenic roots by Agrobacterium rhizogenes-mediated transformation. Plant Cell Tiss Organ Cult. 2020;141:207–16.
Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6(3):325–30.
Dietrich C, Maiss E. Red fluorescent protein DsRed from Discosoma sp. as a reporter protein in higher plants. Biotechniques. 2002;32(2):286–8.
Seki H, Nishizawa T, Tanaka N, Niwa Y, Yoshida S, Muranaka T. Hairy root-activation tagging: a high throughput system for activation tagging in transformed hairy roots. Plant Mol Biol. 2005;59:793–807.
Nishizawa K, Kita Y, Kitayama M, Ishimoto M. A red fluorescent protein, DsRed2, as a visual reporter for transient expression and stable transformation in soybean. Plant Cell Rep. 2006;25(12):1355–61.
Tomilov A, Tomilova N, Yoder J. Agrobacterium tumefaciens and Agrobacterium rhizogenes transformed roots of the parasitic plant Triphysaria versicolor retain parasitic competence. Planta. 2007;225:1059–71.
Lin M, Gresshoff P, Indrasumunar A, Ferguson B. pHairyRed: a novel binary vector containing the DsRed2 reporter gene for visual selection of transgenic hairy roots. Mol Plant. 2011;4:537–45.
Fan Y, Liu J, Lyu S, Wang Q, Yang S, Zhu H. The Soybean Rfg1 gene restricts nodulation by Sinorhizobium fredii USDA193. Front Plant Sci. 2017;8:1548.
Yang S, Hu Y, Cheng Z, Rice JH, Miao L, Ma J, et al. An efficient Agrobacterium-mediated soybean transformation method using green fluorescent protein as a selectable marker. Plant Signal Behav. 2019;14(7):1612682.
Jefferson R, Kavanagh T, Bevan M. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6(13):3901–7.
Mitiouchkina T, Mishin AS, Somermeyer LG, Markina NM, Chepurnyh TV, Guglya EB, et al. Plants with genetically encoded autoluminescence. Nat Biotechnol. 2020. https://doi.org/10.1038/s41587-020-0500-9.
Zuluaga DL, Gonzali S, Loreti E, Pucciariello C, Degl’lnnocenti E, Guidi L, et al. Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants. Funct Plant Biol. 2008;35:606–8.
Li H, Liu N, Shi D, Liu J, Yang W. YAO is a nucleolar WD40-repeat protein critical for embryogenesis and gametogenesis in Arabidopsis. BMC Plant Biol. 2010;10:169.
Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014;14(1):327.
Okamoto S, Yoro E, Suzaki T, Kawaguchi M. Hairy root transformation in Lotus japonicas. BioProtocol. 2013. https://doi.org/10.21769/BioProtoc.795.
Boisson-Dernier A, Chabaud M, Garcia F, Becard G, Rosenberg C, Barker D. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe In. 2001;14:695–700.
Takakazu K, Yasukazu N, Shusei S, Erika A, Tomohiko K, Shigemi S, et al. Complete genome structure of the nitrogen-fixing symbiotic bacterium mesorhizobium loti. DNA Res. 2000;7:331–8.
Cregan PB, Keyser HH, Sadowsky MJ. Host plant effects on nodulation and competitiveness of the Bradyrhizobium japonicum serotype strains constituting serocluster 123. Appl Environ Microb. 1989;55(10):2532–6.
Kereszt A, Li D, Indrasumunar A, Nguyen C, Nontachaiyapoom S, Kinkema M, et al. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat Protoc. 2007;2(4):948–52.
Lü S, Fan Y, Liu L, Liu S, Zhang W, Meng Z. Ectopic expression of TrPI, a Taihangia rupestris (Rosaceae) PI ortholog, causes modifications of vegetative architecture in Arabidopsis. J Plant Physiol. 2010;167(18):1613–21.
Zhang S, Kondorosi É, Kereszt A. An anthocyanin marker for direct visualization of plant transformation and its use to study nitrogen-fixing nodule development. J Plant Res. 2019;132:695–703.
Acknowledgements
We thank Prof. Lijuan Qiu (CAAS, China) for the soybean seeds, Prof. Zhe Yan (Northeast Institute of Geography and Agroecology, CAS, China) for the L. japonicas seeds, Prof. Tianfu Han and Wensheng Hou (CAAS, China) for A. rhizogenes K599 strain, and Prof. Keyuan Tan (South China Agricultural University, China) for USDA110.
Funding
This work was supported by National Natural Science Foundation of China (No. 31271751), and by Natural Science Foundation of Shandong province (Nos. ZR2012CL16 and ZR2010CQ025).
Author information
Authors and Affiliations
Contributions
YF and SL designed the experiments and wrote the paper. YF, XW, HL, SL, LJ, YL, MS, SL and XY performed the work and analyzed data. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors are consent for publications.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Anthocyanin accumulation on transgenic hairy roots containing 35S:: AtMyb75 in various soybean cultivars. Data were obtained by three independent experiment with at least infected 8 seedlings.
Additional file 2: Fig. S1.
Transcription analysis of AtMyb75 in induced soybean hairy roots by RT-PCR. RNAs were extracted from independent hairy roots induced by A. rhizogenes carrying p35AtM75 construct. Gm-Actin (27 cycles) (a) and AtMyb75 (30 cycles) (b) were amplified, respectively. Lane 1–4, non-transgenic white root inoculated with K599-p35AtM75; lane 5, p35AtM75 plasmid; lane 6–11, independent transgenic root with anthocyanin accumulation inoculated with K599-p35AtM75; lane 12, K599; lane 13, ddH2O. M, DL2000 DNA ladder bought from Sangon Biotech (band is 100 bp, 250 bp, 500 bp, 750 bp, 1000 bp, 2000 bp, respectively).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Fan, Y., Wang, X., Li, H. et al. Anthocyanin, a novel and user-friendly reporter for convenient, non-destructive, low cost, directly visual selection of transgenic hairy roots in the study of rhizobia-legume symbiosis. Plant Methods 16, 94 (2020). https://doi.org/10.1186/s13007-020-00638-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13007-020-00638-w