Abstract
Agrobacterium rhizogenes-mediated transformation is widely used in different species with various purposes. The development of composite plants (wild-type shoot with transgenic roots) has been a milestone for functional characterization of genes. Previously, composite plants were generated by two steps from inducing of hairy roots to growing in the growth medium. Hairy roots were induced in an induction medium and the growth of composite plants generated were in another different growth medium. The composite plants produced was subject to transplanting. Here, we describe an improved and optimized protocol for generation of composite plant achieved by one-step in cucumber, which has not been reported previously in living plants. Incubation of explants post inoculation to induce transgenic roots and the growth of rooted explants were in the same medium. The primary root of 5-day-old seedling was excised and the slant cut of residual hypocotyl with 1 cm length was inoculated with A. rhizogenes harboring the desired gene construct followed by directly planted into a pot with wet sterile vermiculite. More than 90% of the infected seedlings can produce positive transgenic root. In addition, we further used the one-step transformation protocol to analyze the function of Arabidopsis YAO promoter. The result indicated that pYAO::GUS was highly conserved expression in whole root and high activity in the root tips. Therefore, a fast, expedient, high efficient, and one-step transformation method of composite cucumber produced is established, which is suitable for promoter functional analysis and other root-related events.
Key message
A highly reliable, facile and one-step generation of composite cucumber plants (with transgenic roots and wild-type shoot) protocol performed effectively in different genotypes cucumber using Agrobacterium rhizogenes-mediated transformation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cucumber (Cucumis sativus L.) is an economically and nutritionally important, and one of the top 5 vegetable crops cultivated worldwide (Liu et al. 2011; Zhao et al. 2019). In 2017, more than 83.8 million tons cucumbers (including gherkins) are globally produced. In 2016, gross production value is 40,229 million US$ (FAO). However, cucumber is high sensitive to low/high temperature, drought/waterlogging and salt stresses that cause significant reductions in yields (Liu et al. 2013, 2010; Ma et al. 2018; Xu et al. 2017, 2018). In addition, a wide range of soilborne diseases (pathogenes), viruses and nematodes are one of the main constraints on cucumber production (Li and Chen 2017; Liang et al. 2019; Liu et al. 2017). Although grafting of cucumber plants onto cucurbitaceous rootstocks is an efficiency way to improve abiotic/biotic stress, this technique is labor-intensive (Xu et al. 2018). Genetic engineering is a powerful and high efficient way to improve cucumber abiotic/biotic stress tolerance.
Cucumber has paid more attention as a model plant for Cucurbitaceae family. Over the past decade, genomics sequencing of cucumber has been completed (Huang et al. 2009). Furthermore, root-specific transcriptome, proteome, EST (expressed sequence tags), large scale cDNA sequencing studies, GWAS (genome-wide association studies) in cucumber have been completed and conducted (Du et al. 2010; Li et al. 2012; Ma et al. 2018; Qi et al. 2012; Wang et al. 2018; Xu et al. 2017; Yuan et al. 2016; Zheng et al. 2019). Efficient transformation methods are the prerequisite for the functional annotations of such numbers of candidate genes and assist the development and improvement of elite cucumber cultivars. However, the functions of most new candidate genes have not been determined due to a lack of efficient transformation method.
Cucumber is one of the recalcitrant species for transformation to obtain the transgenic plants by Agrobacterium tumefaciens-mediated transformation (Nishibayashi et al. 1996). Additionally, it is time-consuming, laborious, and too low efficient to be useful high throughput research. A. rhizogenes-mediated hairy root transformation approach overcomes and complements some shortcomings of the genetic transformation method and offers a convenient alternative and rapid possibility for studying root biology. This transformation method has been widely successfully applied in more than 100 species (Christey 2001; Plasencia et al. 2016; Parks and Yordanov 2020). To date, although widely used in a variety of dicotyledonous species, including several monocots, there has been still no report on generation of composite cucumber plants (with transgenic roots and non-transgenic shoot) in living plants. In previous report, such as in tomato (Ho-Plágaro et al. 2018), soybean (Kereszt et al. 2007), pea (Clemow et al. 2011), composite plants were generated by two steps from inducing of hairy roots to growing in the growth medium. The hairy roots were induced in a medium and then composite plant produced was transplanted to another different growth medium. Here, we reported a fast, simple, high efficient and one-step generation of composite cucumber plants by A. rhizogenes-mediated transformation. Incubation of explants post inoculation to induce transgenic roots and the continuous growth of rooted explants were in the same growth medium. We minimizes the transfer of composite plant to different media and plant manipulation and simplify the method considerably with high transformation efficiency. Furthermore, we applied the transformation method to analyze the expression of Arabidopsis YAO promoter (Li et al. 2010; Yan et al. 2015) to drive the GUS (β-Glucuronidase) reporter gene in cucumber root. The results show that the expression pattern of the YAO in cucumber roots is very similar to that of YAO in Arabidopsis roots. Therefore, a rapid and more simple transformation method of composite cucumber generated was established, which applies for promoter functional analysis and other root biology research.
Materials and methods
Plant materials and growth conditions
Cucumber (Cucumis sativus L.) seeds ‘Chinese long’ inbred line 9930 (genome sequenced) (Huang et al. 2009), and local varieties H3 and Jinyan4 were kept in our lab and surface-sterilized in a 70% ethanol solution for 3 min and then in a containing active sodium hypochlorite 1.2% commercial bleach solution for 5 min, washed five times with sterile distilled water. The sterilized seeds were sown into a pot with sterile wet vermiculite (11.6 cm × 8 cm × 9.5 cm) (watered 0.25 × Gamborg B-5 basal medium) at a depth of 0.5–1 cm. Gamborg B-5 basal medium was purchased in powder form from PhytoTechnology Laboratories (Shawnee Mission, KS, USA). Plants were grown in a growth chamber at 24 ± 2 °C under a 16-h-light/8-h-dark cycle under 80–120 µmol m−2 s−1, approximately 70% relative humidity.
Construction of pRed1305 andYAOpromoter::GUSvectors, andA. rhizogenesstrains
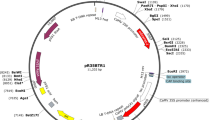
The pRed1305 vector (Fig. 1a) containing a DsRed2 reporter gene driven by constitutively active CaMV 35S promoter was constructed by replacing the Hygromycin Phosphotransferase II resistance marker with the DsRed2 reporter gene in pCAMBIA1305.1. The DsRed2 reporter gene provide the option to identify positive transgenic root from a composite plant based on the fluorescence of DsRed2. To create the pRed1305 vector, the DsRed2 was amplified by PCR using a forward primer RedS1 (5′- CAAGTCGACATGGCCTCCTCCGAGAACGT -3′) and a reverse primer RedS2 (5′- CAAGTCGACCTACAGGAACAGGTGGTGG -3′) using pHairyRed (Addgene Catalog #No.69709) as a template. The forward and reverse primers contain a SalI restriction site, respectively (restriction sites underlined in the primer sequences). The PCR product was digested with SalI and cloned into the dephosphorylated XhoI site of the pCAMBIA1305.1 vector. PCR and enzyme cut analysis were performed to determine DsRed2 inserted into the pCAMBIA1305 vector in the forward orientation. For the pYAO::GUS construct, termed pYGUS1305 (Fig. 1b), a 688 bp YAO promoter fragment (− 587 to + 101 bp) was amplified by PCR using primers YAOF18 (5′- CTGCAGGTACCTCTGAATCGAGCTTTCGGAA -3′) and YAO19R (5′- CTGAACCATGGTCTCTCTCACTCCCTCTTAG -3′) with wild-type Arabidopsis thaliana ecotype Col-0 genomic DNA as template. The forward and reverse primers contain KpnI and NcoI restriction sites, respectively (restriction sites underlined in the primer sequences). The PCR fragment was digested and cloned into the KpnI and NcoI sites of the pCAMBIA1305 vector, where placed YAO promoter to drive the expression of the GUSPlus gene. The pRed1305 and pYGUS1305 vectors were confirmed by sequencing. The binary vector pRed1305 and pYGUS1305 were introduced into A. rhizogenes strain K599 (NCPPB2659; carrying pRi2659 Ri plasmid) (Wang et al. 2016) and ARqual via electroporation, respectively. To get fresh cells, A. rhizogenes strain harboring the desired gene construct (from glycerol stock) was streaked directly onto LB agar plates containing 50 mg/l kanamycin and 50 mg/l streptomycin and incubated for 2 days at 28 °C. A single colony was re-suspended into 1 ml of liquid LB medium (supplemented with antibiotics, 28 °C/200 rpm). After that, the overnight 200 µl of the fresh culture A. rhizogenes was spread onto the surface of LB solid plate (supplemented with antibiotics) and incubated at 28 °C overnight.
One-step A. rhizogenes -mediated root transformation and generation of cucumber composite plants
In the one-step protocol, the primary root was excised from healthy seedlings using a pair of sterile scissors (Fig. 2a). Five-day-old seedlings (the ideal transformation age; the seedlings with just emergence out of vermiculite is marked as 0 day; about 11 days after sowing; the younger seedlings with lower survival rates) with just emergence of the first true leaf (Fig. 2a) were infected on the hypocotyl with A. rhizogenes strain K599 carrying the desired gene construct. The apical portion of the hypocotyl was cut diagonally (0.5 cm cut) with a sterile scalpel in the liquid of A. rhizogenes (OD600 ≈ 0.6–1.0) (Fig. 2b). The slant cut of residual hypocotyl was scraped on the plate grown A. rhizogenes 3–5 times and the bacterial mass was observed in the slant cut (Fig. 2c) followed by directly planted into a pot with wet sterile vermiculite (Fig. 2d). The slanting cut can increase the surface area for optimum bacterial contact. The double inoculation with A. rhizogenes on cut can increase the possibility of bacteria infection. The explants inoculated were cultured under high humidity (covering with a transparent sterile plastic bag or a plastic dome) conditions (Fig. 2d). Key steps of the protocol is shown in Fig. 2. The explants inoculated do not need to water during the following 2 weeks. After 2 weeks, the hairy roots produced. The plastic bag was perforated artificially or adjust ventilation holes of dome to decrease slowly the humidity in order to make the composite plants with transgenic roots can well adapt to environment. After 2–3 days, the plastic bag or dome was removed (Fig. 2e). The composite plants were regularly checked for maintaining water and humidity. Gamborg B-5 basal medium at quarter strength (0.25 ×) was used for watering the seedlings. The induction of hairy roots and growth of composite plant were performed as the previous described in the section of plant materials and growth conditions in this paper.
a Schematic representation of the one-step transformation method of cucumber composite plant produced by cutting the hypocotyl. The primary roots of 5-day-old seedlings were excised using a pair of sterile scissors. b The hypocotyl was cut diagonally (0.5 cm cut) with a sterile scalpel in the liquid of A. rhizogenes. c The cut of residual hypocotyl is scraped on the plate grown A. rhizogenes. d The hairy roots are induced in growth medium (vermiculite) and high humidity is maintained by covering a plastic dome. e The cucumber composite plant with transgenic roots is generated and continue to grow in growth medium without transplanting. The orange roots represent transgenic roots and blue ones represent non-transgenic roots. (Color figure online)
Histochemical GUS staining, DNA extraction and PCR (polymerase chain reaction) analysis
To determine the transgenic and non-transgenic roots, DsRed2 fluorescence and PCR analysis were performed. DsRed2 fluorescence was detected according to Lin et al. (2011). The independent roots transformed were subject to DNA isolation for PCR analysis. Genomic DNA was extracted from each independent root transformed according to the method described previously (McCouch et al. 1988). PCR amplification was carried out in a 20 µl reaction volume containing 2 µl of 10 × PCR buffer, 1.2 µl of dNTPs (10 mmol l−1), 0.3 µl of each primer (10 µmol l−1), 50 ng of DNA template, 1.0 U of Taq DNA polymerase. PCR assay was performed to amplify the DsRed2. The forward primer RedS1 and reverse primer RedS2 were used. The PCR reaction program was conducted as published previously (Lü et al. 2010). In YAO promoter activity analysis, β-Glucuronidase (GUS) activity was assayed by using X-gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronide) as a substrate according to the protocol previously described with some modifications (Jefferson et al. 1987). The transformed roots were stained in GUS staining solution, which contained 100 mM sodium phosphate at pH 7.0, 0.1% Triton X-100, 1 mg/ml X-Gluc, 1 mM potassium ferricyanide, and 1 mM potassium ferrocyanide at 37 °C for 2–10 h. After GUS staining, the samples were rinsed in a 70% ethanol for 10–20 min.
Fluorescent observation
The DsRed fluorescence of cucumber plants was detected using Tanon-5200Multi machine (Tanon Co., Ltd., China) with excitation at 540 nm, emission at 600 nm.
Data analysis
Transformation efficiency was calculated at 21 dpi (days post-inoculation; inoculation date is marked as 0 day). A plant is statistical to be transgenic composite plant as long as it presents at least one transgenic root. Both DsRed2 strong red fluorescence and PCR-positive root was determined as transgenic root. The transformation efficiency is a proportion of numbers of transgenic composite plants compared to total numbers of plants infected. Each experiment was a biological replicate and repeated three times. Data were analyzed using Microsoft office Excel 2016 and Data Processing System (DPS) statistical software. The averages and standard deviations were calculated.
Results
An optimized method for generation of cucumber composite plants by A. rhizogenes -mediated transformation
To obtain cucumber composite plants by A. rhizogenes-mediated root transformation, we first used the most widely applied A. rhizogenes K599 and ARqual strains to infect cucumber. However, ARqual strain are insufficient to infect the cucumber. Transformation efficiency is 0%. By comparison, K599 strains are sufficient to infect cucumber. Hence, K599 was used in all subsequent experiments. In the one-step transformation method by cutting the hypocotyl (Fig. 3a–e), the primary root of seedling was removed and the slant cut of residual hypocotyl was inoculated (Fig. 3a–c) followed by planted in the sterile wet vermiculite (Fig. 3d). All inoculated explants initiated to produce roots at approximately 12 dpi and developed completely rooting process about 21 dpi. The Fig. 3e indicates that an example of cucumber composite plant generated at 21 dpi. The composite plants produced can directly continue to grow in the vermiculite without subjecting to transplanting post-inoculation. In order to determine the optimal transformation protocol, we tested the effects of residual length of the hypocotyl, three different genotypes, and seedling age on transformation efficiency.
Agrobacterium rhizogenes-mediated root transformation of cucumber achieved by one-step. a Five days old healthy seedlings. b The apical portion of the hypocotyl was cut diagonally (0.5 cm cut) in the liquid of K599 harboring the desired gene construct. c A slant cut of the residual hypocotyl was scraped and inoculated on the plate grown A. rhizogenes K599 harboring the desired gene construct. Note the bacterial mass coated in the slant cut. d Explant inoculated was directly planted into a pot with wet sterile vermiculite. e An example of composite plant induced at 21 dpi. Bars = 1 cm
To assess the effect of different length of the residual hypocotyl on transformation efficiency, 0.5 cm, 1.0 cm, 1.5 cm and 2.0 cm residual hypocotyl were maintained. The results indicated that the numbers of positive roots from each seedlings varied from 2.82 ± 1.08 to 3.38 ± 1.57 but there were no significant difference regardless of the length of the residual hypocotyl (Table 1). The 1.0 cm residual hypocotyl displayed the highest transformation efficiency at 95% contrasting to 1.5 cm residual hypocotyl did the lowest at 86.67% (Table 1). To evaluate the effect of different cucumber genotypes on transformation efficiency, three different genotypes were determined. All cucumber genotypes can be infected successfully by K599. While the total numbers of the roots induced were significantly different among different genotypes (Table 2). As for H3, the total numbers of roots induced from the wound and the positive roots were 7.17 ± 2.26 and 3.02 ± 1.83, respectively. The transformation efficiency was the highest at 95% (Table 2). Both the total numbers of roots and positive roots induced in genotype H3 were significantly different from the other two genotypes. The genotype H3 indicated the highest transformation efficiency compared to Jinyan4 and 9930. In contrast, the total numbers of positive roots was the lowest at 1.95 ± 1.17 in genotype 9930 (Table 2). Although both the Jinyan4 and 9930 displayed a little lower transformation efficiency compared with H3, both can reach up to 90% (Table 2). There was no significant difference between the numbers of positive roots between Jinyan4 and 9930 (Table 2). In the three genotypes tested, the percentage of transgenic roots from the total roots per seedling ranged from 32.23 to 43.61% (Table 2). To analyze the effect seedlings age on the transformation efficiency, 5 d-, 7 d-, 9 d-, 11d-old seedlings were used for transformation. The results indicated that the numbers of positive roots did not differ significantly for 5–11 days old seedlings but with some distinct differences on transformation efficiency (Table 3). The numbers of positive roots ranged from 2.77 ± 1.77 to 3.10 ± 2.24. The transformation efficiency decreased with the increase of the seedlings age ranging from 95 to 81.67% (Table 3). In all analysis, the co-transformed roots were identified based on strong DsRed2 fluorescence (Fig. 4) and PCR analysis from each independent transformed roots (Fig. 5). DsRed2 fluorescence was not observed in non-transgenic roots, which is consistent with that of PCR analysis. The PCR detection indicated positive roots, the DsRed2 fluorescence were observed, and vice versa (Figs. 4, 5).
Screening of co-transformed roots expressing the DsRed2 report gene in a composite cucumber plant. a Roots of composite cucumber plant were visualized with a stereomicroscope under bright light at 21 dpi. b Strong fluorescence of DsRed2 were visualized in the co-transformed roots, absence of DsRed2 in non-transformed roots. Bars = 1 cm
Validation of transgenic or non- transgenic roots by PCR. Lane 1, amplification of DsRed2 gene from K599-harboring pRed1305 as a positive control. Lane 2–8, different independent transgenic root from DsRed2-positive root; Lane 9, DsRed2-negative root inoculated with K599 carrying pRed1305. Lane 10, non-transgenic root inoculated with K599 as negative control. Lane 11, ddH2O used as a template.
Application of one-step transformation by cutting the hypocotyl to promoter activity analysis
To evaluate that transgenic roots by A. rhizogenes-mediated transformation were suitable for studying the promoter activities of gene involved in root development, we used the one-step generation of composite plant method to study the promoter activities of YAO (Li et al. 2010) in cucumber roots. We produced independent transgenic root lines expressing the reporter gene GUS under the control of the YAO promoter. pYAO::GUS is expressed in whole root and strong GUS activity was observed in the cucumber root tips (Fig. 6). The expression pattern of YAO in cucumber root is very similar to that of YAO in Arabidopsis (Li et al. 2010). Therefore, we show that the Arabidopsis YAO gene is conserved expressed in cucumber roots.
Discussion
A fast, simple, high efficient and one-step generation of composite cucumber plants with transgenic roots by A. rhizogenes -mediated transformation
It takes 2.5–6 months to obtain cucumber stable genetic transgenic plants by traditional A. tumefaciens-mediated transformation, which is time-consuming, recalcitrant nature and not efficient enough to allow the high-throughput for functional genomic research (Nishibayashi et al. 1996; Zheng et al. 2019). In previous report, to increase the regeneration ratio of transgenic cucumber seedlings, transgenic roots were induced from cucumber hypocotyl using A. rhizogenes in vitro and were used as explants for plantlet regeneration to obtain stable and genetic transgenic plants (Kodama et al. 1993; Trulson et al. 1986). However, it is long transgenic period, labor-intensive and low efficient. The development of composite plants with transgenic roots and wild-type shoot has been a milestone for rapidly assess gene expression and function involved in root biology and improvement of elite species character (Boisson-Dernier et al. 2001). Although transgenic roots were induced from cucumber hypocotyl in vitro using A. rhizogenes (Kodama et al. 1993; Trulson et al. 1986), until this report, there has not been reported on generation of cucumber composite plants in living plants by A. rhizogenes-mediated transformation. Previously, the vitro produced transgenic roots lacking of aerial organs (shoot) removed from tissue cultures media will be stop growing and can not be suitable for analyzing the root to shoot communication in the field of plant hormones, RNAs and proteins because it is absence of systemic signaling patterns. Here, we present a rapid, simple and high efficiency generation of cucumber composite plants with transgenic roots and wild-type shoot by A. rhizogenes-mediated transformation. In our protocol, one-step generation of composite plants can be completed about 1 month with shorter transformation period and higher efficiency.
In previous study, it needs two-steps that the composite plants can grow in the growth medium after inoculation and producing the hairy roots no matter stabbing the hypocotyl inoculation (Kereszt et al. 2007; Meng et al. 2019; Plasencia et al. 2016) or cutting the radical or the primary root inoculation (Boisson-Dernier et al. 2001; Collier et al. 2005). In the most widely inoculation protocol by stabbing the hypocotyl (Kereszt et al. 2007; Meng et al. 2019; Plasencia et al. 2016), the hairy roots were induced in living plant, followed by the primary root of seedlings was cut off and the composite plants were transplanted into the growth medium (Boisson-Dernier et al. 2001; Collier et al. 2005). In the other most widely used inoculation protocol by cutting the primary root or radical, the hairy roots were induced on agar slopes or in rockwool (Fibgro cube) post-inoculated with A. rhizogenes. In the following steps, to detach the hairy roots from the agar or rockwool (Fibgro cube) with taking care without breaking roots and to transplant the composite plants into growth medium (Boisson-Dernier et al. 2001; Collier et al. 2005; Ilina et al. 2012), it is labor-intensive and time-consuming. Ho-Plágaro et al. (2018) made some modifications in the latter protocol. The seedlings was placed between the filter paper to avoid the induced hairy roots growing into the agar. However, no matter the former or the latter modified protocol, two steps are required for the composite plants that can grow in the growth medium. In our protocol, the transgenic roots were induced in the growth medium and do not need to be transplanted and can directly grow in the growth medium. It is only one-step generation of composite plants from inoculation to growing in the growth medium. We minimize the transfer of composite plant to different media and plant manipulation and considerably simplify the method.
In our one-step protocol, for obtaining optimal conditions, we studied the effects of residual length of the hypocotyl, three different genotypes, and seedling age on transformation efficiency. The results indicated that the numbers of positive transgenic roots did not differ significantly for 0.5 cm, 1.0 cm, 1.5 cm and 2 cm length residual hypocotyl and 5 d-, 7 d-, 9 d-, 11 d-old seedlings age (Tables 1, 3). This indicated that hairy roots can be induced on a various length residual hypocotyl and wide span seedling age. However, there are some distinct differences on transformation efficiency among different length residual hypocotyl as well as among different seedlings age (Tables 1, 3). Five days old and seven days old seedlings with 1 cm length residual hypocotyl produced the highest transformation efficiency with 95% (Tables 1, 3). Therefore, for a little shorter transformation period, 5-day-old seedling with 1 cm length residual hypocotyl was suitable for transformation optimal conditions. In all tested genotypes, high transformation efficiency was found ranging from 90 to 95% and the numbers of positive roots indicated a little difference. In some species, such as in chickpea, the transformation efficiency indicated from 23.51 to 61.61% among seven different cultivars (Aggarwal et al. 2018). In soybean, transformation efficiency varied from 30 to 93.3 among seven different genotypes (Cao et al. 2009). Taken together, these data indicated that different genotypes led to different transformation efficiency in different species. Anyway, in cucumber, all genotypes tested can be obtained high transformation efficiency (more than 90%) although H3 genotype is optimal for transformation among the three genotypes tested (Table 2). In addition, in this study, the transformation efficiency is remarkable difference with different A. rhizogenes strains. There were not transformed roots obtained at all with ARqual. In contrast, K599 led to transformation efficiency can reach up to 95%. This is in consistent with previous report, transformation efficiency was variable depending on different A. rhizogenes strains. For example, in pea (Pisum sativum), A. rhizogenes AR12 was less efficient than strain AR1193 (Clemow et al. 2011). In Eucalyptus, ARqual results in transformation efficiency of only 4% and A4 does that of 0%, whereas A4RS ranges from 62% in average to reaching up to 75% depending on the plant material and the infection protocol (Plasencia et al. 2016). In Phaseolus spp., transformation efficiency is 75–90% when cultivars and landraces of Phaseolus spp. were infected with K599, while A4, A15834, A8194 and A2659 are completely ineffective or induce hairy roots only in certain genotypes (Estrada-Navarrete et al. 2007). Therefore, for a given plant species, it needs to screen for a compatible and high efficiency A. rhizogenes strains because co-transformation efficiency were various with different A. rhizogenes strains.
In both previous and our one-step protocol, the hairy roots induced by A. rhizogenes included both transformed and wild type. Non-transformed roots (don’t need to be removed) can serve as a good control for the wild-type phenotype compared with the transformed roots in a composite plant, especially for optimized protocol for functional genomics analyses of candidate genes in root-microbe interactions, such as nematodes, plant-pathogens interaction.
Application of transformation method to study promoter function
So far, A. rhizogenes-mediated hairy root transformation has already been widely applied for many purposes, such as nodulation, mycorrhization, interactions of plants with nematodes or pathogens, drought, salt stress, root nutrient uptake, hormone transport and rooting of recalcitrant species (Cao et al. 2009; Christey 2001; Guillon et al. 2006; Kereszt et al. 2007; Meng et al. 2019). Here, we used the one-step protocol to explore the application of cucumber transgenic roots to analyze promoter function, we conducted Arabidopsis YAO promoter to drive the expression of GUS reporter gene. The result indicated that the Arabidopsis YAO regulatory sequences has shown to drive similar expression patterns of YAO gene in cucumber roots (Li et al. 2010). GUS activity was detected in whole cucumber roots and higher level in the root tips. Previously, higher gene editing efficiency was found by CRISPR/Cas9 system mediated gene knockout using YAO promoter to drive Cas9 protein expression in Arabidopsis and Citrus (Yan et al. 2015; Zhang et al. 2017). It is interesting that the conservation of the expression of Arabidopsis YAO in cucumber roots whether YAO promoter-driven CRISPR/Cas9 system can enhance the gene editing efficiency in cucumber. This need to be further tested. In addition, activities, enhancers, repressors, and the core region of the root specific promoter could be detected easily in the root transformation system (Chen et al. 2014).
Conclusion
We developed a highly reliable and facile one-step generation of composite cucumber plants protocol performed effectively in different genotypes cucumber using A. rhizogenes. It will be easy to transfer a foreign desirable gene into cucumber roots by this method. The fast, simple, high efficient, and reproducible method opens new avenues in desirable agronomical traits improvement of cucumbers roots and elucidate the functions of candidate genes involved in root biology such as the interactions of plants with nematodes or root specific pathogens, nutrient/toxin uptake or remediation from soil.
References
Aggarwal P, Nag P, Choudhary P, Chakraborty N, Chakraborty S (2018) Genotype-independent Agrobacterium rhizogenes-mediated root transformation of chickpea: a rapid and efficient method for reverse genetics studies. Plant Method 14:55
Boisson-Dernier A, Chabaud M, Garcia F, Becard G, Rosenberg C, Barker D (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. MPMI 14:695–700
Cao D, Hou W, Song S, Sun H, Wu C, Gao Y, Han T (2009) Assessment of conditions affecting Agrobacterium rhizogenes-mediated transformation of soybean. Plant Cell Tissue Org Cult 96:45–52
Chen L, Jiang B, Wu C, Sun S, Hou W, Han T (2014) GmPRP2 promoter drives root-preferential expression in transgenic Arabidopsis and soybean hairy roots. BMC Plant Biol 14:245
Christey M (2001) Use of ri-mediated transformation for production of transgenic plants . In Vitro Cell Dev Biol Plant 37:687–700
Clemow S, Clairmont L, Madsen L, Guinel F (2011) Reproducible hairy root transformation and spot-inoculation methods to study root symbioses of pea. Plant Method 7:46
Collier R, Fuchs B, Walter N, Lutke W, Taylor C (2005) Ex vitro composite plants: an inexpensive, rapid method for root biology. Plant J 43:449–457
Du C, Fan H, Guo S, Tezuka T, Li J (2010) Proteomic analysis of cucumber seedling roots subjected to salt stress. Phytochemistry 71:1450–1459
Estrada-Navarrete G et al (2007) Fast, efficient and reproducible genetic transformation of Phaseolus spp. by Agrobacterium rhizogenes. Nat Protoc 2:1819–1824. https://doi.org/10.1038/nprot.2007.259
FAO. http://faostat.fao.org. Accessed 20 Oct 2019
Guillon S, Trémouillaux-Guiller J, Pati P, Rideau M, Gantet P (2006) Hairy root research: recent scenario and exciting prospects. Curr Opin Plant Biol 9:341–346
Ho-Plágaro T, Huertas R, Tamayo-Navarrete M, Ocampo J, García-Garrido J (2018) An improved method for Agrobacterium rhizogenes-mediated transformation of tomato suitable for the study of arbuscular mycorrhizal symbiosis. Plant Method 14:34
Huang S et al (2009) The genome of the cucumber Cucumis sativus L. Nat Genet 41:1275–1281. https://doi.org/10.1038/ng.475
Ilina E, Logachov A, Laplaze L, Demchenko N, Pawlowski K, Demchenko K (2012) Composite Cucurbita pepo plants with transgenic roots as a tool to study root development. Ann Bot 110:479–489
Jefferson R, Kavanagh T, Bevan M (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Kereszt A, Li D, Indrasumunar A, Nguyen C, Nontachaiyapoom S, Kinkema M, Gresshoff P (2007) Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat Protoc 2:948–952
Kodama H, Irifune K, Kamada H, Morikawa H (1993) Transgenic roots produced by introducing Ri-rol genes into cucumber cotyledons by particle bombardment. Transgenic Res 2:147–152
Li H, Liu N, Shi D, Liu J, Yang W (2010) YAO is a nucleolar WD40-repeat protein critical for embryogenesis and gametogenesis in Arabidopsis. BMC Plant Biol 10:169
Li J, Sun J, Yang Y, Guo S, Glick B (2012) Identification of hypoxic-responsive proteins in cucumber roots using a proteomic approach. Plant Physiol Biochem 51:74–80
Li X, Chen S (2017) Screening and identification of cucumber germplasm and rootstock resistance against the root-knot nematode (Meloidogyne incognita). Genet Mol Res 16:gmr16029383. https://doi.org/10.4238/gmr16029383
Liang C, Liu H, Hao J, Li J, Luo L (2019) Expression profiling and regulatory network of cucumber microRNAs and their putative target genes in response to cucumber green mottle mosaic virus infection. Arch Virol 164:1121–1134
Lin M, Gresshoff P, Indrasumunar A, Ferguson B (2011) pHairyRed: A novel binary vector containing the DsRed2 reporter gene for visual selection of transgenic hairy roots. Mol Plant 4(3):537–545
Liu L, Duan L, Zhang J, Mi G, Zhang X, Zhang Z, Ren H (2013) Arabidopsis LOS5 gene enhances chilling and salt stress tolerance in Cucumber. J Integr Agric 12:825–834
Liu L, Duan L, Zhang J, Zhang Z, Mi G, Ren H (2010) Cucumber (Cucumis sativus L.) over-expressing cold-induced transcriptome regulator ICE1 exhibits changed morphological characters and enhances chilling tolerance. Sci Hortic 124:29–33
Liu SQ, Liu XH, Jiang LW (2011) Genome-wide identification, phylogeny and expression analysis of the lipoxygenase gene family in cucumber. Genet Mol Res 10:2613–2636. https://doi.org/10.4238/2011.October.25.9
Liu Y, Chen L, Wu G, Feng H, Zhang G (2017) Identification of root-secreted compounds involved in the communication between cucumber, the beneficial Bacillus amyloliquefaciens, and the soil-borne pathogen Fusarium oxysporum. Mol Plant Microb Interact 30:53–62
Lü S, Fan Y, Liu L, Liu S, Zhang W, Meng Z (2010) Ectopic expression of TrPI, a Taihangia rupestris (Rosaceae) PI ortholog, causes modifications of vegetative architecture in Arabidopsis. J Plant Physiol 167:1613–1621
Ma J et al (2018) Transcriptome analysis of cucumber roots reveals key cold-resistance genes induced by AM fungi Plant. Mol Biol Rep 36:135–148
McCouch S, Kochert Q, Yu Z, Wang Z, Khush G, Coffman W, Tanksley S (1988) Molecular mapping of rice chromosome. Theor Appl Genet 26:815–829
Meng D et al (2019) Development of an efficient root transgenic system for pigeon pea and its application to other important economically plants. Plant Biotechnol J. https://doi.org/10.1111/pbi.13101
Nishibayashi S, Hayakawa T, Nakajima T, Suzuki M, Kaneko H (1996) CMV protection in transgenic cucumber plants with an introduced CMV-O cp gene. Theor Appl Genet 93:672–678
Parks T, Yordanov YS (2020) Composite plants for a composite plant: an efficient protocol for root studies in the sunflower using composite plants approach. Plant Cell Tissue Organ Cult. https://doi.org/10.1007/s11240-019-01760-x
Plasencia A, Soler M, Dupas A et al (2016) Eucalyptus hairy roots, a fast, efficient and versatile tool to explore function and expression of genes involved in wood formation. Plant Biotechnol J 14:1381–1393. https://doi.org/10.1111/pbi.12502
Qi X, Xu X, Lin X, Zhang W, Chen X (2012) Identification of differentially expressed genes in cucumber (Cucumis sativus L.) root under waterlogging stress by digital gene expression profile. Genomics 99:160–168
Trulson A, Simpson R, Shahin E (1986) Transformation of cucumber (Cucumis sativus L.) plants with Agrobacterium rhizogenes. Theor Appl Genet 73:11–15
Wang S, Song Y, Xiang T et al (2016) Transgenesis of Agrobacterium rhizogenes K599 orf3 into plant alters plant phenotype to dwarf and branch. Plant Cell Tissue Organ Cult 127:207–215. https://doi.org/10.1007/s11240-016-1043-0
Wang X et al (2018) The USDA cucumber (Cucumis sativus L.) collection: genetic diversity, population structure, genome-wide association studies, and core collection development. Hortic Res 5:64. https://doi.org/10.1038/s41438-018-0080-8
Xu X, Chen M, Ji J, Xu Q, Qi X, Chen X (2017) Comparative RNA-seq based transcriptome profiling of waterlogging response in cucumber hypocotyls reveals novel insights into the de novo adventitious root primordia initiation. BMC Plant Biol 17:129
Xu Y, Yuan Y, Du N, Wang Y, Shu S, Sun J, Guo S (2018) Proteomic analysis of heat stress resistance of cucumber leaves when grafted onto Momordica rootstock. Hortic Res 5:53
Yan L, Wei S, Wu Y, Hu R, Li H, Yang W, Xie Q (2015) High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol Plant 8:1820–1823
Yuan Y, Zhong M, Shu S, Du N, Sun J, Guo S (2016) Proteomic and physiological analyses reveal putrescine responses in roots of cucumber stressed by NaCl. Front Plant Sci 7:1035
Zhang F, LeBlanc C, Irish V, Jacob Y (2017) Rapid and efficiency CRISPR/Cas9 gene editing in Citrus using the YAO promoter. Plant Cell Rep 36:1883–1887
Zhao J et al (2019) A functional allele of CsFUL1 regulates fruit length through inhibiting CsSUP and auxin transport in cucumber. Plant Cell. https://doi.org/10.1105/tpc.18.00905
Zheng Y et al (2019) Cucurbit Genomics Database (CuGenDB): a central portal for comparative and functional genomics of cucurbit crops. Nucleic Acids Res 47:D1128–D1136. https://doi.org/10.1093/nar/gky944
Acknowledgements
We are grateful to prof. Tianfu Han and Wensheng Hou (Chinese Academy Agriculture of Sciences) for kindly providing the A. rhizogenesis K599 strain. We are also thankful to prof. Qi Xie (Institute of Genetics and Developmental Biology, Chinese Academy of Science) for kindly providing p1300-pYAO-Cas9 vector. This work was funded by grants from National Natural Science Foundation of China (Grant Number 31271751) and by Natural Science Foundation of Shandong province (Grant Numbers ZR2012CL16 and ZR2010CQ025). The funding agencies were not involved in the designing of the study and collection, analysis and interpretation of the data, and in the writing of the manuscript.
Author information
Authors and Affiliations
Contributions
YF and SL designed the experiments and wrote the paper. YF, FX, HZ, XL, XY, KW, and XS carried out the experiments. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Editorial Responsibility: Maria Margarida Oliveira.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fan, Y., Xu, F., Zhou, H. et al. A fast, simple, high efficient and one-step generation of composite cucumber plants with transgenic roots by Agrobacterium rhizogenes-mediated transformation. Plant Cell Tiss Organ Cult 141, 207–216 (2020). https://doi.org/10.1007/s11240-020-01781-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-020-01781-x