Abstract
Background
The Anopheles gambiae sensu lato (s.l.) and Anopheles funestus s.l. species complexes contain the most important malaria vectors in Africa. Within the An. funestus group of at least 11 African species, the vector status of all but the nominal species An. funestus appears poorly investigated, although evidence exists that Anopheles rivulorum and Anopheles vaneedeni may play minor roles. A new species, An. rivulorum-like, was described from Burkina Faso in 2000 and subsequently also found in Cameroon and Zambia. This is the first paper reporting the presence of this species in South Africa, thereby significantly extending its known range.
Methods
Mosquitoes were collected using dry-ice baited net traps and CDC light traps in the Kruger National Park, South Africa. Sixty-four An. funestus s.l. among an overall 844 mosquitoes were captured and identified to species level using the polymerase chain reaction assay. All samples were also analysed for the presence of Plasmodium falciparum circumsporozoite protein using the enzyme-linked-immunosorbent assay.
Results
Four members of the An. funestus group were identified: An. rivulorum-like (n = 49), An. rivulorum (n = 11), Anopheles parensis (n = 2) and Anopheles leesoni (n = 1). One mosquito could not be identified. No evidence of P. falciparum was detected in any of the specimens.
Conclusion
This is the first report of An. rivulorum-like south of Zambia, and essentially extends the range of this species from West Africa down to South Africa. Given the continental-scale drive towards malaria elimination and the challenges faced by countries in the elimination phase to understand and resolve residual transmission, efforts should be directed towards determining the largely unknown malaria vector potential of members of the An. funestus group and other potential secondary vectors.
Similar content being viewed by others
Background
The most important and widespread vectors of malaria in Africa are members of two species complexes: Anopheles gambiae sensu lato (s.l.) with at least eight sibling species [1], and Anopheles funestus s.l. with 11 African species [2, 3]. Adult members of these two complexes are difficult or impossible to separate morphologically [4,5,6], requiring molecular techniques for reliable identification [1, 7,8,9]. Species within both complexes vary greatly in feeding behaviour, host preferences and in particular their efficiency as vectors of malaria [4,5,6, 10, 11].
Within the An. funestus group, An. funestus sensu stricto (s.s.) is widely acknowledged as one of the three most efficient and important vectors of malaria in Africa [4, 5, 12]. As for other members of the An. funestus group, some reports indicate that An. rivulorum may be involved in malaria transmission in some situations [13,14,15], and Plasmodium falciparum has been reported from Anopheles parensis and Anopheles leesoni [14]. Anopheles vaneedeni has been experimentally infected with P. falciparum [16], and P. falciparum has recently been isolated in natural populations of this species in South Africa [11]. No reports of any involvement in malaria transmission for the remaining members of the An. funestus group were found. Because of the close morphological similarity but very different malaria transmission capacities of these species, it is important to know which occur in a particular area as this influences malaria vector control decisions and operations, vital commitments in all African countries where financial and related malaria control resources are limited.
The Afrotropical members of the An. funestus group can be categorized into three subgroups (Table 1), of which the Rivulorum Subgroup with four species is of relevance here. Aside from Anopheles rivulorum, Anopheles brucei and Anopheles fuscivenosus, this Subgroup also includes An. rivulorum-like that was first described from Burkina Faso as a cryptic but unnamed taxon [17], subsequently called “An. rivulorum-like” by Cohuet et al. [9] based on specimens caught in Cameroon. Later, Norris and Norris [18] reported An. rivulorum-like also present in Zambia. Its presence has not been recorded elsewhere.
In this paper, findings indicating populations of An. rivulorum-like in two localities within the Kruger National Park, north-eastern South Africa, are presented, thereby expanding the known range of this species from West Africa (Burkina Faso, Cameroon) and Zambia down to South Africa, the southern limit of malaria transmission in Africa.
Methods
Study area
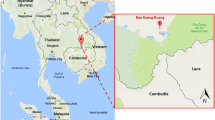
Mosquito collections were undertaken in March 2015 in the Kruger National Park as part of a broader study on mosquito distribution and abundance in South Africa. The Kruger National Park (KNP) is located in the north-eastern corner of South Africa, bordering Mozambique to the east and Zimbabwe to the north. The specific localities where traps were placed within this ca 19,000 km2 National Park are the Shingwedzi River within 20 km west and east of Shingwedzi Camp (S23° 06.656′, E31° 27.419′), and in the mostly dry bed of the Nwaswitsontso River within 500 metres of the Tshokwane Picnic Site (S24° 47.121′, E31° 51.283′) (Fig. 1). Shingwedzi is in the northern part of the KNP, within the Limpopo Province of South Africa, and Tshokwane in the Mpumalanga Province, both these Provinces being malaria endemic. All sites where anophelines were collected had standing pools of water in the river-bed with an abundant growth of aquatic plants floating on the surface.
Mosquito collections
Mosquito net traps [19] and CDC light traps [20] baited with dry-ice were used to collect adult mosquitoes. All Anopheles mosquitoes were identified using morphological keys [5], and members of the An. gambiae and An. funestus groups were individually placed in silica gel tubes for subsequent PCR identification. Sixty-four An. funestus s.l. were captured, representing 7.6% of the total 844 mosquitoes collected.
Identification of Anopheles funestus members
Genomic DNA from Anopheles mosquitoes were individually extracted from two legs/two wings in 30 µl sodium chloride-Tris–EDTA (STE) buffer (1 M NaCl, 1 M Tris–HCl and 0.5 M EDTA). Specimens were ground in STE buffer and incubated for 12 min at 94 °C. The extracted DNA was assayed by PCR using species-specific primers to separate six members of the An. funestus group including An. rivulorum-like [8, 9]. Anopheles funestus species-specific PCR was done according to the protocol described by Koekemoer et al. [8]. Positive and negative controls were added with the unidentified Anopheles specimens in each PCR reaction. Positive controls included the following species of the An. funestus group: An. funestus, An. leesoni, An. parensis, An. rivulorum and An. vaneedeni. The negative controls included a PCR mastermix without template as well as a DNA extraction negative control (DNA extraction performed without mosquito sample).
Following amplification, 10 µl of the PCR products were fragment-size separated through a 2.5% agarose gel stained with ethidium bromide and were visualized on a UV trans-illuminator. To confirm the identity of the unexpected An. rivulorum-like specimens, amplicons from these specimens were excised from the gel, purified using Wizard®SV (Promega) and sequenced using the ABI 3130 Genetic Analyser by Macrogen Inc.
Confirmation of An. rivulorum-like species identification
DNA was extracted from three specimens that were suspected to be An. rivulorum-like, using prepGEM® DNA Extraction Kits (ZyGEM™). The internal transcribed spacer region 2 (ITS2), which is the noncoding region between the 5.8S and 28S coding region of the specimens was amplified by PCR according to the protocol by Koekemoer et al. [8].
Subsequently, the PCR products of the ITS2 region from the three specimens were purified and sequenced by Macrogen Inc. The chromatograms of the sequences were analysed by using BioEdit version 7.2.5 [21]. The resulting sequences of the 3 specimens were compared to each other by using the Muscle multiple sequence alignment [22] and a consensus sequence was established. Subsequently, the Emboss Needle pairwise sequence alignment tool [23] was used to compare the consensus sequence with An. rivulorum-like sequences with the following GenBank accession numbers: KR014818 [24], JN994147 [18] and AF210725 [17] as well as an An. rivulorum sequence GenBank accession number JN994148 [18].
Plasmodium falciparum detection
The head and thorax of each female mosquito was subjected to indirect enzyme-linked immunosorbent assay (ELISA) for presence of P. falciparum circumsporozoite protein (CSP) using monoclonal antibodies 2A10 as described by Wirtz et al. [25]. One positive control and seven negative controls were added to each microtitre plate. The positive control consisted of a synthetic peptide standardized against the human malaria parasite, P. falciparum. Negative controls were unfed An. funestus s.s. from laboratory colonies maintained by the Vector Control Reference Laboratory, National Institute for Communicable Diseases, Johannesburg. Absorbance was measured at 405 nm using a microtitre plate reader. The cut-off value for positive specimens was taken as twice the mean value of the negative controls.
Results
Of the 64 Anopheles funestus s.l. identified morphologically, 63 were identified to species level by PCR as An. rivulorum-like (n = 49), An. rivulorum (n = 11), An. parensis (n = 2) and An. leesoni (n = 1). One sample failed to amplify, which may be due to human error, DNA degradation or morphological miss-identification. The PCR of the An. rivulorum-like specimens produced a diagnostic 313 bp fragment [9] (Fig. 2). The species-specific PCR was repeated twice to confirm the results.
An. funestus group PCR confirmed that the amplified PCR fragment of the Shingwedzi specimens corresponds to the An. rivulorum-like size fragment of 313 bp. Lane 1 and 11: 100 bp DNA ladder; lane 2: An. funestus positive control; lane 3: An. leesoni positive control; lane 4: An. parensis positive control; lane 5: An. rivulorum positive control; lane 6: An. vaneedeni positive control; lane 7: negative control; lanes 8–10: three specimens from Shingwedzi
The ITS2 region of three suspected An. rivulorum-like specimens was amplified by PCR and produced an amplicon of ~ 550 bp (Fig. 3) which corresponds to the ITS2 region of An. rivulorum. Sequencing analysis of the ITS2 region from the specimens confirmed that the three specimens had a 100% identity. The established consensus sequence for the An. rivulorum-like from South Africa is:
PCR of the ITS2 region of members of the An. funestus group and three examples of the Shingwedzi (South Africa) specimens. PCR amplification from the specimens produced an amplicon of ~ 550 bp, which is similar to the An. rivulorum size fragment. Lanes 1 and 11: 100 bp DNA ladder; lane 2: An. funestus; lane 3: An. leesoni; lane 4: An. parensis; lane 5: An. rivulorum; lane 6: An. vaneedeni; lane 7: negative control; lanes 8–10: three specimens from Shingwedzi
>An. rivulorum-like (South Africa) GTTTAAACTCGGCCGATGCACACATTCTTGAGTGCCTACCAAATCCTTGATACACAAACACCTGACTACAGTGGTGCACGCGTGCAGCGAACTAAGCACTATGGCGAGACCCACGTCTAGTGTCGCTGAACCACACGCGCCTGCCCACTGTGCATAATGGCGTGCTCGGGAAGTAAAATTCTCGGGGGCGCTGAAGAGCGATGAGAGCATGGGGGCGGTACTCTGTTGCTGCCGGATCCCCCACTCCACGGGAGGCGGATGGTGCGTGTCTAGTTGCGTGTTGCGAACTCTGGCAGGACGTCCTGACAGCCCCGACAGCCCTGCATCAGGATTGTCGGCCTACTGTATCAGGGCCAAACGGCCGGCCAGGTCGCGTAATGCTCGCAGCTTAGACGTGCCACTCCGTCGCCATCGCACGAAAAACCGTAG.
There was a 99.5 and 99.3% sequence identity between the ITS2 region of the 3 specimens and the An. rivulorum-like from Eastern Zambia (GenBank accession number KR0148180) [24] and the An. rivulorum-like from the Southern province Zambia (GenBank accession number JN994147) [18]. The ITS2 region of the three specimens had a sequence identity of 97% with the ITS2 region from the An. rivulorum-like from Burkina Faso (GenBank accession number AF210725) [17]. The homology between the ITS2 region of the three specimens and that of An. rivulorum s.s. (GenBank accession number JN994148) [18] was 80%.
Females comprised 100% of the 64 An. funestus group captured, and ELISA results for P. falciparum were negative for all.
Discussion
The primary vectors of malaria and their geographic distribution are well known, and control efforts directed at such primary vectors has achieved major successes in malaria reduction over the past decade [26]. The real challenge arises in the malaria elimination stages when traditional or standard methods of control need supplementation to address the remaining residual malaria transmission. In countries in the malaria elimination stage, such as South Africa, the causes of at least a portion of such residual malaria are not always clear, resulting in inexplicable small outbreaks or perpetuation of low levels of transmission which pose a threat to elimination targets and no small source of frustration. Secondary, or minor vectors, may be a contributing factor to such residual malaria transmission, especially when little is known regarding the presence or status of such secondary vectors, for example the recent finding in South Africa of An. vaneedeni naturally infected with P. falciparum [11]. It is for this reason that control programmes need to look wider during such critical phases of the final push to eliminate local transmission. Anopheles rivulorum is known to have the potential for low levels of transmission [13,14,15, 24], but nothing appears to be known regarding the vector status of An. rivulorum-like. This paper shows that An. rivulorum-like is fairly common in two areas of the Kruger National Park, north-eastern South Africa, where non-targeted, random collections of mosquitoes were made for non-malaria surveillance purposes.
The ITS2 region of the An. rivulorum-like from South Africa was more closely related to An. rivulorum-like from Zambia (99%) than the ITS2 region of An. rivulorum-like from Burkina Faso (97%). This divergence between the Zambian and Burkina Faso material was also noted by Norris and Norris [18] and they suggested it could be due to geographic variants. However, An. longipalpis type C and An. vaneedeni are seen as two distinct species, yet they show a 97.5% sequence similarity within the ITS2 region [27]. Without additional taxonomic, chromosomal and mating studies, it is impossible to predict if An. rivulorum-like from Burkina Faso and southern Africa are different species or mere geographical variants. With that being said, the sequence identity between the ITS2 region of the South African specimens and that of An. rivulorum was 80%, signifying that these specimens are more closely related to An. rivulorum-like than to An. rivulorum.
Greater effort should be directed towards establishing the potential role of this species in malaria transmission, with particular regard to residual malaria. The negative results for P. falciparum from female An. rivulorum-like reported in this paper are not surprising given that the collections were done inside the KNP, a strictly managed nature reserve. The KNP attracts almost exclusively middle to higher income tourists that are very unlikely to have infective gametocytes, therefore not representative of the situation in the malaria-endemic areas immediately adjoining this nature reserve, on both the South African and Mozambican sides. Interestingly, light trap collections by one of the authors (LB) in October 2017 along the Nwanedzi River (S22° 21.248′ E30° 35.434′) in Vhembe District, Limpopo Province, some 120 km northwest of Shingwedzi, yielded 52 An. funestus group members of which 48 were An. rivulorum, two An. leesoni and three failed PCR testing. This absence of An. rivulorum-like in a geographically close and environmentally similar setting, suggests either microhabitat differences in breeding preferences or seasonal variations in abundance.
In summary, An. funestus s.s. is known as one of the three most widespread and efficient vectors of malaria in Africa, with strong habits of endophagy, endophily and anthropophily. Anopheles rivulorum has been implicated in malaria transmission or found to harbour P. falciparum parasites in Kenya [15], Tanzania [13, 14] and Zambia [24]. Anopheles vaneedeni has been experimentally infected with Plasmodium in the laboratory [16] and recently found infected in nature [11]. As for the other members of the An. funestus group, there is one report [14] that possibly implicates An. leesoni and An. parensis in hosting P. falciparum, but not much appears to have been done to assess their role as secondary or minor vectors of malaria. The taxonomy/classification of An. rivulorum-like remains to be studied, that will require future comparative larval morphologies and whole genome sequencing. Formal naming of An. rivulorum-like is also required.
Conclusion
The finding of An. rivulorum-like in South Africa now extends the range of this species to various points-of-presence straddling virtually the entire sub-Saharan region of Africa, with confirmed identifications in Burkina Faso, Cameroon, Zambia and South Africa.
With several African countries in the malaria elimination stage, and hopefully many more to reach this stage in the years ahead, the contribution of secondary malaria vectors becomes important. Little is known regarding the vector status of most members of the An. funestus group, other than An. funestus s.s. itself. At least one member, An. rivulorum, is known to host P. falciparum, and An. vaneedeni has been recently implicated. Two other species An. parensis and An. leesoni may also play a role as minor vectors. Anopheles rivulorum-like appears to be a fairly common species in parts of South Africa, a country which has ambitious targets for malaria elimination. The malaria vector capacity of An. rivulorum-like therefore needs to be assessed—as indeed do all other potential secondary vectors—especially in relation to the often unclear factors contributing to residual malaria transmission in a malaria elimination context.
Abbreviations
- CDC:
-
Centers for Disease Control and Prevention
- ELISA:
-
enzyme linked immunosorbent assay
- ITS2:
-
internal transcribed spacer region 2
- KNP:
-
Kruger National Park
- PCR:
-
polymerase chain reaction
- rDNE:
-
ribosomal deoxyribonucleic acid
- s.l.:
-
sensu lato
- s.s.:
-
sensu stricto
References
Coetzee M, Hunt RH, Wilkerson R, Della Torre A, Coulibaly MB, Besansky NJ. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa. 2013;3619:246–74.
Harbach RE. The classification of genus Anopheles (Diptera: Culicidae): a working hypothesis of phylogenetic relationships. Bull Entomol Res. 2004;94:537–53.
Coetzee M, Koekemoer LL. Molecular systematics and insecticide resistance in the major African malaria vector, Anopheles funestus. Annu Rev Entomol. 2013;58:393–412.
Gillies MT, De Meillon B. The Anophelinae of Africa south of the Sahara, vol. 54. Johannesburg: Publications of the South African Institute for Medical Research; 1968.
Gillies MT, Coetzee M. A supplement to the Anophelinae of Africa south of the Sahara, vol. 55. Johannesburg: Publications of the South African Institute for Medical Research; 1987.
Spillings BL, Brooke BD, Koekemoer LL, Chiphwanya J, Coetzee M, Hunt R. A new species concealed by Anopheles funestus Giles, a major malaria vector in Africa. Am J Trop Med Hyg. 2009;81:510–5.
Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49:520–9.
Koekemoer LL, Kamau L, Hunt R, Coetzee M. A cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: Culicidae) group. Am J Trop Med Hyg. 2002;6:804–11.
Cohuet A, Simard F, Toto JC, Kengne P, Coetzee M, Fontenille D. Species identification within the Anopheles funestus group of malaria vectors in Cameroon and evidence for a new species. Am J Trop Med Hyg. 2003;69:200–5.
Vezenegho SB, Chiphwanya J, Hunt RH, Coetzee M, Bass C, Koekemoer LL. Characterization of the Anopheles funestus group including Anopheles funestus-like from northern Malawi. Trans R Soc Trop Med Hyg. 2013;107:753–62.
Burke A, Dandalo L, Munhenga G, Dahan Y, Mbokazi F, Coetzee M, et al. A new malaria vector mosquito in South Africa. Sci Rep. 2017;7:43779.
Sinka ME, Bangs MJ, Manguin S, Coetzee M, Mbogo CM, Hemingway J. The dominant Anopheles vectors of human malaria in Africa, Europe and the Middle East: occurrence data, distribution maps and bionoimic précis. Parasit Vectors. 2010;3:117.
Wilkes TJ, Matola YG, Charlwood JD. Anopheles rivulorum, a vector of malaria in Africa. Med Vet Entomol. 1996;10:108–10.
Temu EA, Minjas JN, Tuno N, Kawada H, Takagi M. Identification of four members of the Anopheles funestus (Diptera: Culicidae) group and their role in Plasmodium falciparum transmission in Bagamoyo coastal Tanzania. Acta Trop. 2007;102:119–25.
Kawada H, Dida GO, Sonye G, Njenga SM, Mwandawiro C. Reconsideration of Anopheles rivulorum as a vector of Plasmodium falciparum in Western Kenya: some evidence from biting time, blood preference, sporozoite positive rate, and pyrethroid resistance. Parasit Vectors. 2012;5:230.
De Meillon B, Van Eeden GJ, Coetzee L, Coetzee M, Meiswinkel R, Du Toit CLN, et al. Observations on a species of the Anopheles funestus subgroup, a suspected exophilic vector of malaria parasites in northeastern Transvaal, South Africa. Mosq News. 1977;37:657–61.
Hackett BJ, Gimnig J, Guelbeogo W, Costantini C, Koekemoer LL, Coetzee M, et al. Ribosomal DNA internal transcribed spacer (ITS2) sequences differentiate Anopheles funestus and An. rivulorum and uncover a cryptic taxon. Insect Mol Biol. 2000;9:369–74.
Norris LC, Norris DE. Phylogeny of anopheline (Diptera: Culicidae) species in southern Africa, based on nuclear and mitochondrial genes. J Vector Ecol. 2015;40:16–27.
Service MW. Mosquito ecology. London: Applied Science Publishers; 1976.
Sudia WD, Chamberlain RW. Battery-operated light trap, an improved model. J Am Mosq Control Assoc. 1988;4:536–8.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8.
https://www.ebi.ac.uk/Tools/msa/muscle/. Accessed Jan 2018.
https://www.ebi.ac.uk/Tools/psa/emboss_needle/nucleotide.html. Accessed Jan 2018.
Lobo NF, Laurent BS, Sikaala CH, Hamainza B, Chanda J, Chinula D, et al. Unexpected diversity of Anopheles species in Eastern Zambia: implications for evaluating vector behaviour and interventions using molecular tools. Sci Rep. 2015;5:17952.
Wirtz RA, Zavala F, Charoenvit Y, Campbell GH, Burkot TR, Schneider I, et al. Comparative testing of monoclonal antibodies against Plasmodium falciparum sporozoites for ELISA development. Bull World Health Organ. 1987;65:39–45.
WHO. World malaria report 2016. Geneva: World Health Organization; 2016.
Koekemoer LL, Misiani EA, Hunt RH, Kent RJ, Norris DE, Coetzee M. Cryptic species within Anopheles longipalpis from southern Africa and phylogenetic comparison with members of the An. funestus group. Bull Entomol Res. 2009;99:41–9.
Dia I, Geolbeogo MW, Ayala D. Advances and perspectives in the study of the malaria mosquito Anopheles funestus. In: Manguin S, editor. Anopheles mosquitoes—new insights into malaria vectors, vol. 7. Rijeka: InTech; 2013. p. 197–220. https://doi.org/10.5772/55389.
Kyalo D, Amratia P, Mundia CW, Mbogo CM, Coetzee M, Snow RW. A geo-coded inventory of anophelines in the Afrotropical Region south of the Sahara: 1898–2016. Wellcome Open Res. 2017;2:57. https://doi.org/10.12688/wellcomeopenres.12187.1.
Authors’ contributions
Mosquito collections were done by LB and AJC. Initial PCR and ELISA assays were done by JM with follow-on additional PCR assays by LK and YD-M. The initial draft of the paper was written jointly by JM and LB. Subsequent additions and editorial comments were made by JM, AJC, YD-M, LK, MC, LB. All authors read and approved the final manuscript.
Acknowledgements
LB and AJC thank the Carnegie Foundation for an African Diaspora Fellowship grant. We are grateful to Dr Danny Govender, Sharon Thompson and SANParks staff for generous logistical assistance, and the UP ISMC and MRC Collaborating Centre for Malaria Research of the University of Pretoria for library and other institutional support during the writing of this publication.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
This paper was partly based on a literature survey. We searched PubMed (https://www.ncbi.nlm.nih.gov/nlmcatalog/journals), Google Scholar, Google search and websites of relevant international organization. Search terms initially were “funestus AND Group” or “rivulorum-like”. Abstracts were screened for relevance and appropriate papers then read in full. These primary papers led to other references by way of publications quoted in their texts. The data supporting the conclusions of this article are included within the article.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
AJC was the recipient of a Carnegie African Diaspora Fellowship Grant. MC and LLK are supported by a DST/NRF South African Research Chairs Initiative Grant.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Mouatcho, J., Cornel, A.J., Dahan-Moss, Y. et al. Detection of Anopheles rivulorum-like, a member of the Anopheles funestus group, in South Africa. Malar J 17, 195 (2018). https://doi.org/10.1186/s12936-018-2353-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-018-2353-y