Abstract
Background
Although a high genetic diversity of Plasmodium spp. circulating in great apes has been revealed recently due to non-invasive methods enabling detection in faecal samples, little is known about the actual mechanisms underlying the presence of Plasmodium DNA in faeces. Great apes are commonly infected by strongylid nematodes, including hookworms, which cause intestinal bleeding. The impact of strongylid infections on the detection of Plasmodium DNA in faeces was assessed in wild, western, lowland gorillas from Dzanga Sangha Protected Areas, Central African Republic and eastern chimpanzees from Kalinzu Forest Reserve, Uganda.
Methods
Fifty-one faecal samples from 22 habituated gorillas and 74 samples from 15 habituated chimpanzees were analysed using Cytochrome-b PCR assay and coprological methods.
Results
Overall, 26.4% of the analysed samples were positive for both Plasmodium spp. and strongylids. However, the results showed no significant impact of intensity of infections of strongylids on detection of Plasmodium DNA in gorilla and chimpanzee faeces.
Conclusion
Bleeding caused by strongylid nematode Necator spp. cannot explain the presence of Plasmodium DNA in ape faeces.
Similar content being viewed by others
Background
Helminths are among the most ubiquitous parasite infections in primates and the distribution of helminth parasites is prominent in the areas where malarial infection occurs, resulting in high rates of co-infection [1]. A plethora of studies in humans have studied malaria and helminth co-infections with conflicting results [2]. In contrast, analogous studies in non-human primates (NHPs) have been restricted by the limitation of diagnosing malaria from blood and the obvious challenges associated with invasive sampling of wild primates. However, recent advances in molecular diagnostics enable malaria detection from non-invasively obtained faecal samples in primates [3, 4] and have paved the way for such non-invasive studies in the wild. Previously, studies in NHPs focused on the genetic diversity of Plasmodium spp. [3] and/or ecological aspects [5], no study has investigated co-infections with malaria and other pathogens.
Despite the pivotal role of non-invasive faecal sampling, little is yet known about the causal mechanism that lies behind the presence of ape Plasmodium DNA in faeces. Recently, a single experimental study demonstrated that the pre-erythrocytic stage of the malaria parasite can be detected in mouse faeces, and suggested that parasite DNA gets into the faeces via the bile duct [6]. However, it remains unclear if a similar mechanism applies to Plasmodium DNA entering ape faeces or if under natural conditions other mechanisms might be involved in the shedding of Plasmodium stages into the gastro-intestinal tract. Great apes are commonly infected by strongylid nematodes [7, 8] including hookworms [9]. Hookworms, Ancylostoma duodenale and Necator spp., can cause significant blood loss when the adult parasites attach themselves to the intestinal mucosa and while feeding [10], and thus their presence and consecutive bleeding in the intestine might be a plausible explanation for the detection of Plasmodium DNA in faeces.
Methods
Since 2007 we have been systematically surveying parasite infections of humans, western lowland gorillas and other wildlife inhabiting Dzanga Sangha Protected Areas (DSPA), Central African Republic (CAR), as part of an ongoing health-monitoring programme [5, 9]. Previous research has shown that both gorillas in DSPA and chimpanzees in Kalinzu Forest Reserve (KFR), Uganda were infected with strongylid nematodes [9], (Hasegawa and Pafčo, unpublished data) and with malaria parasites Plasmodium spp. [3, 5]. In this study, co-infections with malaria and strongylid nematodes were explored in both gorillas and chimpanzees. The hypothesis was that a higher infection intensity of strongylid nematodes can result in higher detection rates of Plasmodium spp. in ape faeces.
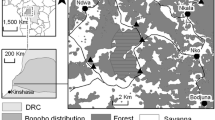
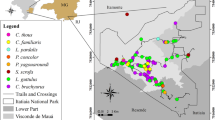
In 2012 51 faecal samples from nine (n = 30 samples) and 13 (n = 21 samples) individuals were collected from two habituated gorilla groups (Makumba, Mayele) in DSPA, CAR. For further description of the field site and studied gorilla groups see Mapua et al. [5]. In 2014 74 faecal samples from 15 individuals from one habituated chimpanzee group (M group) were collected in KFR, Uganda. For a more detailed description of the study sites refer to Yasuoka et al. [11].
Most of the animals were sampled at least twice; however, six gorillas were sampled only once. The faecal samples were fixed immediately into RNAlater™ (Ambion, Austin, TX, USA) for molecular detection of Plasmodium DNA and into 10% formalin for coprological analyses. Formalin-fixed samples were stored at ambient temperature, and RNAlater-fixed samples were stored at 4 °C in a refrigerator at the base camp. The samples were subsequently shipped to the Czech Republic and those stored in RNAlater were kept at −20 °C until DNA extraction. The methods for molecular detection of malaria, including DNA extraction, PCR, sequencing, and phylogenetic analyses are described in detail in Mapua et al. [5].
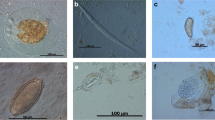
For initial coprological examination Sheather’s flotation with modified sugar solution (s.g. 1.33) [12] and the sedimentation method [13] were used. To assess infection intensity of strongylid nematodes the number of eggs per gram of sediment (EPG) were quantified directly from the sediment [14]. The exact determination of strongylids to genus or even species level is mostly unreliable based on egg morphology alone. Previous morphological and molecular analyses of filariform larvae obtained from coprocultures from studied groups of both gorillas and chimpanzees showed the thin-walled strongylid eggs belonged to either the genus Oesophagostomum or Necator (Fig. 1: Hasegawa and Pafčo, unpublished data, [9]). Of the two, only Necator spp. are known to cause intestinal bleeding, however the thin-walled strongylid eggs were categorized as ‘strongylid nematodes/strongylids’ and counts may have included eggs of both Oesophagostomum and Necator. Morphological analyses of L3 larvae from pilot sets of coprocultures showed that all coprocultures contained Necator spp., however only 54 and 29% for gorillas and chimpanzees, respectively, contained larvae of Oesophagostomum spp. (Hasegawa and Pafčo, unpublished data). Mammomonogamus sp., the only strongylid eggs which can be easily distinguished by microscopy, can be also found in faecal samples of gorillas in DSPA [14]. Mammomonogamus spp. eggs were excluded from ‘strongylid nematodes/strongylids’ egg counts as adults of Mammomonogamus spp. occur in the respiratory tract of the host not in the gastro-intestinal [15].
Prevalence was defined as the number of positive samples for both Plasmodium DNA and strongylid eggs divided by the total of samples tested. Statistical analyses were performed using R software v 3.1.3; R Development Core Team 2015 [16, 17]. The effect of strongylid infection intensity on malaria detection in faeces was tested using generalized linear mixed effects models (GLMMs), with the presence of Plasmodium spp. as a binomial response variable (positive/negative for Plasmodium spp. DNA in faeces) and individual identity and host species (gorilla and chimpanzees) included as random factors. GLMMs were fitted by maximum likelihood with the binomial error distribution and implemented using the glmer function of the R package lme4.
Results
Results of the molecular analyses of Plasmodium spp. infecting western lowland gorillas in DSPA were reported in detail in Mapua et al. [5]. In eastern chimpanzees the host specific strains of Plasmodium reichenowi, Plasmodium billbrayi and Plasmodium billcollinsi were reported [18]. All examined chimpanzees and gorillas were positive for strongylid nematodes except three gorilla samples (see Appendix). Of the total 74 chimpanzees samples analysed, 12.16% (9 samples) were positive for both Plasmodium spp. and strongylid nematodes, in gorillas it was 47.06% (24 samples) from total of 51 samples. The mean intensity of strongylid infection in gorillas was 82.86 EPG (range 4–428) for malaria positive and 77.97 EPG (range 0–500) for malaria negative samples, in chimpanzees it was 263.13 EPG (range 51–370) for malaria positive and 281.90 EPG (range 13–1478) for malaria negative samples (Fig. 2). There was no significant effect of infection intensity of strongylid nematodes on detection rate in faeces for Plasmodium spp. DNA (z = −0.679, P = 0.497).
Discussion
The results did not support the hypothesis that bleeding caused by strongylid nematode Necator spp. can explain the presence of Plasmodium DNA in western lowland gorilla and eastern chimpanzee faeces. The magnitude of Necator spp. pathogenesis and blood shedding is highly dependent on the number of adult worms, host immunity and concurrent infections with other nematodes [19]. To properly estimate infection intensity of a particular gastro-intestinal parasite species, invasive collection of adult parasites from the intestines of infected hosts is required, which is indeed impossible in wild endangered animals like great apes, with exception of necropsies. Due to individual and temporal variation in egg output, it is questionable whether the number of parasite eggs shed in faeces is linearly correlated with infection intensity [20]. However, several studies have found a linear relationship between faecal number of eggs and adult worm number in hosts [e.g., 20] and thus number of eggs has been used as a proxy for intensity of parasite infection also in non-invasive studies of non-human primates [21].
The results can be affected by the fact that it is impossible to distinguish eggs of Necator spp. from other strongylids, which are not associated with intestinal bleeding (as Oesophagostomum spp.) with the exception of Mammomonogamus sp. However, preliminary results indicate that Necator spp. are more prevalent in both studied ape populations and although Oesophagostomum infections are usually not associated with intestinal bleeding in humans, there is a report of rectal bleeding due to Oesophagostomum brumpti in a human [22]. However, in general, data about clinical manifestations and pathogenesis of any strongylid infections in wild non-human primates are scarce [23].
Taking an alternative perspective, Plasmodium pathogenesis itself may facilitate shedding of the parasites into the gastrointestinal tract [6]. It is known that the spleen plays a central role in phagocytosis of different pathogens including blood-stages of Plasmodium [24]. The liver is known as the site of pre-erythrocytic development of Plasmodium parasites and also participates in degradation and removal of the infected red blood cells from the circulation [24]. Recently, an experimental study on rodent malaria showed that Plasmodium DNA is detectable in the liver and gall bladder, as well as in faeces [6]. It was suggested that Plasmodium DNA derived from pre-erythrocytic stage parasites were released into the gastro-intestinal tract via the bile duct. Given that this experimental study was based on the inoculation of a high number of sporozoites and detection of Plasmodium DNA shortly after infection, it is questionable whether a similar phenomenon occurs in different hosts that are exposed to long-term infections with low parasitaemia levels under natural conditions. The inability to obtain wild ape blood samples, as well as predicting when the animals get infected, make difficult to suggest which Plasmodium stages could be detected in the ape faeces, and if Plasmodium DNA detected in this study underwent similar mechanism.
Conclusions
Although no significant impact of strongylid infections on detection of Plasmodium DNA in faeces of wild apes was found, further studies are warranted to better understand the relationship between hookworms and malaria infections. Faecal and blood samples should be examined from sub-clinical malaria-positive humans with and without strongylid hookworms or the use an experimentally co-infected primate model to understand the possible role of hookworm infections in the detection of Plasmodium DNA in faeces.
References
Petney TN, Andrews RH. Multiparasite communities in animals and humans: frequency, structure and pathogenic significance. Int J Parasitol. 1998;28:377–93.
Mwangi TW, Bethony JM, Brooker S. Malaria and helminth interactions in humans: an epidemiological viewpoint. Ann Trop Med Parasitol. 2006;100:551–70.
Liu W, Li Y, Learn GH, Rudicell RS, Robertson JD, Keele BF, et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;476:420–5.
Jirků M, Pomajbíková K, Petrželková KJ, Hůzová Z, Modrý D, Lukeš J. Detection of Plasmodium spp. in human feces. Emerg Infect Dis. 2012;18:634–6.
Mapua MI, Qablan MA, Pomajbíková K, Petrželková KJ, Rádrová J, Votýpka J, et al. Ecology of malaria infections in western lowland gorillas inhabiting Dzanga Sangha Protected Areas, Central African Republic. Parasitology. 2015;142:890–900.
Abkallo HM, Liu W, Hokama S, Ferreira PE, Nakazawa S, Maeno Y, et al. DNA from pre-erythrocytic stage malaria parasites is detectable by PCR in the feces and blood of hosts. Int J Parasitol. 2014;144:467–73.
Huffman MA, Gotoh S, Turner L, Yoshida K. Seasonal trends in intestinal nematode infection and medicinal plant use among chimpanzees in the Mahale Mountains, Tanzania. Primates. 1997;38:111–25.
Makouloutou P, Mbehang Nguema PP, Fujita S, Takenoshita Y, Hasegawa H, Yanagida T, et al. Prevalence and genetic diversity of Oesophagostomum stephanostomum in wild lowland gorillas at Moukalaba-Doudou National Park, Gabon. Helminthologia. 2014;51:83–93.
Hasegawa H, Modrý D, Kitagawa M, Shutt KA, Todd A, Kalousová B, et al. Humans and great apes cohabiting the forest ecosystem in Central African Republic harbour the same hookworms. PLoS Negl Trop Dis. 2014;8:e2715.
Hotez PJ, Brooker S, Bethony JM, Bottazzi ME, Loukas A, Xiao SH. Hookworm infection. N Engl J Med. 2004;351:799–807.
Yasuoka H, Kimura D, Hashimoto C, Furuichi T. Quantitative assessment of livelihoods around great ape reserves: cases in Luo Scientific Reserve, DR Congo, and Kalinzu Forest Reserve, Uganda. Afr Study Monogr. 2012;43:137–59.
Sheather AL. The detection of intestinal protozoa and mange parasites by flotation technique. J Comp Pathol. 1923;36:266–75.
Blagg W, Schloegel EL, Mansour NS, Khalaf GI. A new concentration technique for the demonstration of protozoa and helminth eggs in feces. Am J Trop Med Hyg. 1955;4:23–8.
Kalousová B. Gastrointestinal parasites of wild western lowland gorillas (Gorilla gorilla gorilla) in Dzanga Sector of Dzanga-Ndoki National Park, Central African Republic. M.S. Thesis, Masaryk University; 2013. p. 39–41.
Nosanchuk JS, Wade SE, Landolf M. Case-report of and description of parasites in Mamomonogamus laryngeus (human synga-mosis) infection. J Clin Microbiol. 1995;33:998–1000.
R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: The R Foundation for Statistical Computing. 2011. ISBN 3-900051-07-0. http://www.R-project.org/.
Mapua MI, Petrželková KJ, Burgunder J, Dadáková E, Brožová K, Hrazdilová K, et al. A comparative molecular survey of malaria prevalence among Eastern chimpanzee populations in Issa Valley (Tanzania) and Kalinzu (Uganda). Malar J. 2016;15:423.
Robertson LJ, Crompton DW, Sanjur D, Nesheim MC. Haemoglobin concentrations and concomitant infections of hookworm and Trichuris trichiura in Panamanian primary school children. Trans R Soc Trop Med Hyg. 1992;86:654–6.
Cabaret J, Gasnier N, Jacquiet P. Faecal egg counts are representative of digestive-tract strongyle worm burdens in sheep and goats. Parasite. 1998;5:137–42.
Seivwright LJ, Redpath SM, Mougeot F, Watt L, Hudson PJ. Faecal egg counts provide a reliable measure of Trichostrongylus tenuis intensities in free-living red grouse Lagopus lagopus scoticus. J Helminthol. 2004;78:69–76.
Leoutsakos B, Agnadi N, Kolisiatis S. Rectal bleeding due to Oesophagostomum brumpti: report of a case. Dis Colon Rectum. 1977;20:632–4.
Krief S, Jamart A, Mahé S, Leendertz FH, Mätz-Rensing K, Crespeau F, et al. Clinical and pathologic manifestation of oesophagostomosis in African great apes: does self-medication in wild apes influence disease progression? J Med Primatol. 2008;37:188–95.
Buffet PA, Safeukui I, Deplaine G, Brousse V, Prendki V, Thellier M, et al. The pathogenesis of Plasmodium falciparum malaria in humans: insights from splenic physiology. Blood. 2011;117:381–92.
Helmby H. Pathology exacerbates malaria-induced liver gastrointestinal nematode infection. J Immunol. 2009;182:5663–71.
Authors’ contributions
MIM, BK and KJP designed, and KJP and DM supervised the study. KJP and JB completed the sampling material and fieldwork; AT and CH mentored the field survey. MIM, BK, JB, and IP conducted the laboratory examination and molecular analyses of the samples. MIM, BK, MAQ, JB, and KJP analysed the data. MIM, BK and KJP wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank the government of the Central African Republic and the World Wildlife Fund for granting permission to conduct our research in the Central African Republic; the Ministère de l’Education Nationale, de l’Alphabétisation, de l’Enseignement Supérieur, et de la Recherche for providing research permits; and the Primate Habituation Programme.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets analysed during the current study are available in the Table 1 and from the corresponding author on reasonable request.
Ethics approval and consent to participate
Samples were collected non-invasively and thus no observable distress was caused to the animals. All procedures performed in the study were in accordance with the ethical standards of the institutional and/or national research committee (Ethical Commission of the Biology Centre of the Academy of Sciences of the Czech Republic).
Funding
This work was funded by Czech Science Foundation (15-05180S), by the project CEITEC-Central European Institute of Technology (CZ.1.05/1.100/02.0068) from European Regional Development Fund, Internal Grant Agency of University of Veterinary and Pharmaceutical Sciences Brno (90/2014/FVL), co-financed from European Social Fund and state budget of the Czech Republic (project OPVK CZ.1.07/2.3.00/20.0300) and finally supported by the Institute of Vertebrate Biology Academy of Sciences of the Czech Republic (RVO: 68081766).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
See Table 1.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Mapua, M.I., Pafčo, B., Burgunder, J. et al. No impact of strongylid infections on the detection of Plasmodium spp. in faeces of western lowland gorillas and eastern chimpanzees. Malar J 16, 175 (2017). https://doi.org/10.1186/s12936-017-1822-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-017-1822-z