Abstract
Background
The aberrant regulation of MALAT1 has been indicated to be involved in various carcinogenic pathways contributing to the tumourigenesis and progression of cancers. The current meta-analysis summarized the research advances of MALAT1 functions and analyzed its prognostic value among multiple types of cancers.
Methods
Eligible studies were identified through retrieving the PubMed, Web of Science, and CNKI databases, up to Mar 1, 2018. 28 studies of 5436 patients and 36 studies of 3325 patients were enrolled in the meta-analysis to evaluate the association of MALAT1 expression with survival outcomes and clinical parameters.
Results
The results demonstrated that over-expression of MALAT1 may predict lymph node metastasis (pooled OR = 2.335, 95% CI 1.606–3.395, P = 0.000) and distant metastasis (pooled OR = 2.456, 95% CI 1.407–4.286, P = 0.002). Moreover, MALAT1 was also related with tumour size (pooled OR = 1.875, 95% CI 1.257–2.795, P = 0.002) and TNM stage (pooled OR = 2.034, 95% CI 1.111–3.724, P = 0.021). Additionally, elevated MALAT1 expression could predict poor OS (pooled HR = 2.298, 95% CI 1.953–2.704, P = 0.000), DFS (pooled HR = 2.036, 95% CI 1.240–3.342, P = 0.005), RFS (pooled HR = 2.491, 95% CI 1.505–4.123, P = 0.000), DSS (pooled HR = 2.098, 95% CI 1.372–3.211, P = 0.001) and PFS (pooled HR = 1.842, 95% CI 1.138–2.983, P = 0.013) in multivariate model. Importantly, subgroup analyses disclosed that increased MALAT1 expression had a poor OS among different cancer types (Estrogen-dependent cancer: pooled HR = 2.656, 95% CI 1.560–4.523; urological cancer: pooled HR = 1.952, 95% CI 1.189–3.204; glioma: pooled HR = 2.315, 95% CI 1.643–3.263; digestive cancer: pooled HR = 2.451, 95% CI 1.862–3.227).
Conclusions
The present findings demonstrated that MALAT1 may be a novel biomarker for predicting survival outcome, lymph node metastasis and distant metastasis.
Similar content being viewed by others
Background
Long non-coding RNAs (lncRNAs) as genomic “dark matters” have been disclosed to be closely related to the development of cancer [1,2,3], which lead to the alteration of oncogenic phenotypes including cell proliferation, differentiation, metastasis, apoptosis and invasion [3,4,5,6]. Based on the current emerging evidence, cancer-related lncRNAs may be candidate biomarkers for affording precise diagnosis, appraisal of personalized prognosis, evaluation of targeted therapy and prediction of lymph node metastasis, distant metastasis as well as tumour differentiation [7,8,9].
The metastasis-associated lung adenocarcinoma transcript 1 (MAlAT1) is mapped to human chromosome 11q13 [10, 11]. Differentiating from other members of lncRNAs family, MALAT1 is a broadly expressed and evolutionarily conserved lncRNA with length of more than 8000 nt. Ji et al. initially discovered that MALAT1, a metastasis-associated gene, may be used to identify early-stage NSCLC patients that are at high risk to develop deterioration with metastasis [12]. Evidence for the carcinogenic roles of MALAT1 has gradually emerged from previous fundamental and clinical studies. For example, MALAT1 silencing might impede proliferation, migration, and invasion of triple-negative breast cancer (TNBC) cell by reversely mediating MiR-129-5p [13]. MALAT1 acts as a competitive endogenous RNA (ceRNA) to regulate ZEB1 expression by sponging miR-143-3p, whereas miR-143-3p inhibitor partially impaired the effect of MALAT1 on hepatocellular carcinoma (HCC) cells, and the inhibition of MALAT1 also might inhabit proliferation and invasion of HCC cells [14]. Moreover, activation of p53 may be due to depletion of MALAT1, which may result in cell cycle defects that are sensitive to p53 levels [15]. MALAT1 expression might be regulated by DNA methylation in lung cancer by evaluating methylation status of the CpG island at the MALAT1 promoter [16]. Furthermore, up-regulated MALAT1 promotes cell metastasis by activating the Wnt/β-catenin signaling pathway to promote EMT of bladder cancer cells [17, 18]. Transition of oncogenic phenotypes caused by MALAT1 have also been observed in cervical cancer [19], gastric cancer [20], prostate cancer [21]. Taken together, emerging evidence manifested that dysregulated MATAT1 is closely related to the development of various types of cancer.
For the recent decade, increasing studies have demonstrated the influence of MALAT1 expression on clinicopathological parameters and prognostic outcomes among diffident types of cancer, including digestive cancers [20, 22,23,24,25,26], gliomas [27, 28], estrogen-dependent cancers [29,30,31], urological cancers [32,33,34] and other cancers [12, 35]. However, these published studies have emerged the inconsistent and controversial conclusions [22, 32, 34, 36]. Herein, we conducted a systematic review and meta-analysis to elucidate the relationship of MALAT1 with prognosis or clinical features and generalized its tumorigenicity among different cancers.
Materials and methods
Literature search
Eligible records were systematically retrieved in three authoritative databases including PubMed, Web of Science, and CNKI databases up to March 1, 2018 to obtain relevant articles regarding prognostic and clinicopathological outcomes of MALAT1 among malignant cancers, with the following keywords including “MALAT1 expression and (outcome or prognosis or prognostic or mortality or survival) and (cancer OR carcinoma OR tumor OR malignancy OR neoplasm OR lymphoma OR leukemia)”. Besides, the references lists of included studies were retrieved to guarantee that all qualified studies contained in the pooling analysis.
Study selection and data extraction
Data extraction of each qualified articles was as follows: first author, year, country, ethnicity, type of cancers, follow-up (months), detection method, sample size, survival outcome and the corresponding HR and 95% CI and other data for clinical parameters. Eligible articles need to meet the following criteria: (a) studies with cancers diagnosed by pathological and histological confirmation; (b) studies with the survival outcomes such as “overall survival, “disease-free survival”, “recurrence-free survival”, “disease-specific survival”, “progression-free survival”, recurrence and mortality, and other clinical parameters such as lymph node metastasis, distant metastasis, differentiation/histological grade, tumor size and TNM stage; (c) original studies detected MALAT1 expression in tissue or plasma; (d) studies did explicitly provide HR and 95% CI. However, ineligible articles were excluded on the basis of the following criteria: (a) studies focused on other lncRNAs, diagnosis, polymorphism, case reports, reviews and meta-analyses; (b) studies did not provide available data; (c) studies only with mechanisms of MALAT1 and other genes; (d) animal studies of MALAT1 and other lncRNAs; (e) duplicated published reports, articles or data.
Quality assessment
Two investigators individually assessed the quality of all included studies according to the Newcastle–Ottawa Scale (NOS), and the scale totally comprises subject selection, comparability of study groups as well as ascertainment of survival outcomes. Articles with NOS ≥ 6 scores were regard as high-quality studies.
Statistical analysis
Cochran’s Q and I2 tests were applied to find the heterogeneity across studies. Hazard ratios (HRs), odds ratios (ORs) and their 95% confidence intervals (95% CIs) were calculated by using a random effect model when I2 > 50% and the corresponding P value < 0.05. Otherwise, a fixed effect model was used to estimate the pooled results. Subgroup analysis were further performed to find the source of heterogeneity. Each single study on the overall effect of the stability of the pooled results was estimated by performing sensitivity analyses. Egger’s test and Begg’s funnel plot were applied to identify publication bias. All calculated results of the meta-analysis were performed by using Stata 11 software. A P value < 0.05 was consistently regarded as statistical significance.
Results
Identification of the included studies
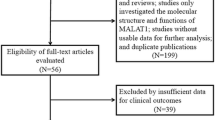
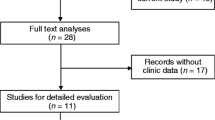
In the study, the detailed selection process of all 48 included articles presented in Fig. 1. A total of 5436 patients from 28 articles covering 54 cohort studies were included to evaluate prognostic value (presented in Additional file 1: Table S1, Additional file 2: Table S2). Of 54 studies with survival outcomes including OS, DFS, RFS, PFS and DSS, 25 studies from 19 articles [20, 22,23,24, 27, 29, 32, 34, 37,38,39,40,41,42,43,44,45,46,47,48] in univariate analysis, 29 studies from 21 articles [14, 23, 27,28,29, 32, 34, 35, 37, 39, 41,42,43,44,45,46,47, 49,50,51,52] in multivariate analysis. Additionally, 3325 patients from 36 articles [13, 17, 20, 23,24,25,26,27, 31,32,33,34, 37, 42,43,44, 46,47,48,49,50, 53,54,55,56,57,58,59,60,61,62,63,64,65,66,67] with clinical parameters including age, gender, lymph node metastasis (LNM), distant metastasis, differentiation, tumor size and TNM stage were enrolled in the study (data shown in Additional file 3: Table S3). The study contains four cancer types including digestive cancers with gastric cancer (GC), gallbladder cancer (GBC), esophageal cancer (EC), pancreatic duct adenocarcinoma (PDAC), esophageal squamous cell carcinoma (ESCC), hepatocellular carcinoma (HCC) and colorectal cancer (CRC); gliomas with glioblastoma, glioma and glioblastoma multiforme (GBM); estrogen-dependent cancers with cervical cancer (CC), epithelial ovarian cancer (EOC) and breast cancer (BC); and urological cancers with urothelial carcinoma (UC), bladder cancer and clear cell renal cell carcinoma (ccRCC). MALAT expression was detected by quantitative real time PCR (qRT-PCR) and in situ hybridization (ISH).
Association of MALAT1 with clinicopathological parameters
As presented in Table 1, the significant association of MALAT1 expression with patients’ age or gender did not existed (age: P = 0.823 and gender: P = 0.080). The increased expression level of MALAT1 was significantly associated with lymph node metastasis (pooled OR = 2.335, 95% CI 1.606–3.395, P = 0.000), tumour size (pooled OR = 1.875, 95% CI 1.257–2.795, P = 0.002), distant metastasis (pooled OR = 2.456, 95% CI 1.407–4.286, P = 0.002) and TNM stage (pooled OR = 2.034, 95% CI 1.111–3.724, P = 0.021). Moreover, subgroup analysis of cancer type presented that patients with MALAT1 over-expression had higher risk of lymph node metastasis and distant metastasis (shown in Fig. 2a, b).
Association of MALAT1 with survival outcomes
A total of 15 eligible studies with 1869 cases focused on evaluating the association between MALAT1 expression and overall survival in univariate model (Table 2). Overall, patients with elevated expression of MALAT1 had a poor OS in univariate (pooled HR = 2.296, 95% CI 1.716–3.072, P = 0.000) analysis with heterogeneity (I2 = 67.6%). Unlike subgroup of univariate analysis, almost all analytical results of multivariate subgroup had no significant heterogeneity (I2 < 50%). 18 studies comprising 1891 patients reported the relationship of MALAT1 expression with OS in multivariate analysis. Overexpression of MALAT1 had a higher risk of poor OS (pooled HR = 2.298, 95% CI 1.953–2.704, P = 0.000, I2 = 17.2%). MALAT1 high expression was also indicated to predict poor OS among different cancer types (Estrogen-dependent cancer: pooled HR = 2.656, 95% CI = 1.560–4.523; Urological cancer: pooled HR = 1.952, 95% CI 1.189–3.204; Glioma: pooled HR = 2.315, 95% CI 1.643–3.263; Digestive cancer: pooled HR = 2.451, 95% CI 1.862–3.227) (data shown in Table 2 and Fig. 3a). Subgroup analysis of sample size presented in Fig. 3b.
Table 3 presented that patients with MALAT1 over-expression had shorter DFS (pooled HR = 2.036, 95% CI 1.240–3.342, P = 0.005), RFS (pooled HR = 2.491, 95% CI 1.505–4.123, P = 0.000), DSS (pooled HR = 2.098, 95% CI 1.372–3.211, P = 0.001) and PFS (pooled HR = 1.842, 95% CI 1.138–2.983, P = 0.013) than those with low MALAT1 expression in a fixed-effect model (shown in Table 3 and Fig. 4).
Publication bias and sensitivity analysis
Egger’s test and Begg’s funnel plot were applied to examine publication bias. Egger’s test revealed absence of publication bias for OS in univariate (T = 1.47, P = 0.164) and multivariate (T = 1.55, P = 0.141) analyses. The symmetrical funnel plot of OS in multivariate model was presented in Fig. 5. The Egger’s tests and funnel plots of DFS, DSS and RFS also showed no publication bias. Furthermore, no significant publication bias was observed in clinicopathological parameters except for LNM (P = 0.041) and differentiation (P = 0.003). The studies of Droop et al. [36] significantly influenced the pooled results of OS and DFS according to sensitivity analysis, which indicated that the studies might explain the main source of heterogeneity across studies. Reanalyzed sensitivity analysis identified that the results of the study remained stability and robustness after getting rid of the studies of Droop et al.
Carcinogenic mechanisms of MALAT1 among various cancers
To further explore the association of MALAT1 with the development of cancer, we summarized the effects of MALAT1 silencing on the malignant phenotype and its molecular mechanisms presented in Table 4. Additionally, MALAT1 possesses a variety of molecular functions including promotion of EMT [68], transcriptional dysregulation, pre-mRNA alternative splicing, ceRNA role [69], epigenetic alteration and transition of cell phenotype via different signaling pathways covering P13k/Akt [62], Wnt [18] and ERK/MAPK [70] pathways. Taken together, MALAT1 might promote carcinogenesis by exerting its molecular function to regulate the expression of related genes and activate the oncogenic signaling pathway [16, 46, 52, 64, 68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83].
Discussion
MALAT1 also known as NEAT2 (nuclear-enriched abundant transcript 2), is located in human chromosome 11q13. Unlike most of lncRNAs, MALAT1 is particularly abundant, highly conserved and ubiquitously expressed in multiple types of cancer. MALAT1 was originally discovered to predict metastasis and survival of non-small cell lung cancer [12]. Recently, increasing evidence provided that MALAT1 play a pivotal role in promoting proliferation, migration, metastasis and invasion of tumor cell. MALAT1, a multi-functional lncRNA, might involve in alternative splicing of pre-mRNA, transcriptional and post-transcriptional regulation via interacting with the relevant gene in carcinogenic pathways [84, 85]. Firstly, MALAT1, a novel transcript, may recruit a set of members of SR protein (serine/arginie riched protein) family, such as SRSF1, SRSF2, and SRSF3, and act as a “molecular sponge” to regulate SR protein activity, ultimately leading to alternative splicing of pre-mRNA [86]. Secondly, MALAT1 involvement in transcriptional dysregulation was supported by previous evidence, such as colocalization of serine-2 phosphorylated RNA polymerase II in nuclear speckle compartment, the interaction of unmethylated Pc2 with theTUG1, overlapping the histone H3K36me2 peaks and the recruitment of Sp1 on LTBP3 promoter. For example, MALAT1 could interact with unmethylated Pc2 in the nuclear speckles, and regulate the localization of the Pc2, together with theTUG1, whereas methylated Pc2 generally exists in other nuclear bodies [87]. MALAT1 also inclined to cooperate with the 3′ end of the gene body, overlapping the histone H3K36me2 peaks, a biomarker of active transcriptional elongation [10]. Thirdly, the mechanisms of the post-transcriptional regulation of MALAT1 mainly contains alternative splicing, protein activities and ceRNAs. For example, MALAT1, as a ceRNA, could reciprocally interacts with microRNAs (miR-205, miR-1297, miR-217 and miR-155), ultimately contributing to cell phenotypic changes such as invasiveness, metastasis, proliferation, migration and apoptosis [52, 72, 77]. Besides, MALAT1 might influence carcinogenesis of cancers by activating Wnt/β-catenin, ERK/MAPK and PI3K/AKT pathways, which simultaneous activation of the oncogenic pathways might bring out highly carcinogenic effects [88]. For example, knockdown of MALAT1 could induce the EMT by regulating transcriptional factor snail and activating the PI3K/AKT and Wnt pathways [31, 33, 80]. Furthermore, upregulated MALAT1 could promote EMT-mediated cell migration and metastasis of various malignant cancers since its inhibition impairs the effect of TGF-β-induced EMT by suppressor of zeste 12 (suz12) [17]. Hence, emerging studies have implied that MALAT1 could serve as a potential prognostic biomarker for cancer patients on the basis of the complicated mechanisms of MALAT1 among multiple types of cancer.
Previous published articles reported that lncRNAs including MALAT1 are effective predictors of survival outcomes [89, 90]. However, MALAT1 on the influence of prognostic outcome is still controversial. Therefore, we combined the published studies to evaluate the prognostic and clinical value of MALAT1 among different types of cancer. The meta-analysis is firstly to investigate the relationship between MALAT1 expression and prognosis of glioma as well as estrogen-dependent cancer by pooling eligible studies in multivariate model, which is different from previous meta-analyses. The pooled results of multivariate model may be closer to revealing the authentic relationship of MALAT1 expression with different types of cancers since the adjusted confounding factors involved in several clinical variables as confounders including LNM, differentiation, distant metastasis and other factors. This study also firstly analyzed the association of MALAT1 with tumour differentiation, distant metastasis, TNM stage and tumour size.
The results of the study identified that patients with high expression of MALAT1 have a poor OS in univariate and multivariate models. Moreover, over-expression of MALAT1 may be an unfavorable biomarker of DFS, RFS, DSS and PFS. The results also indicated that the adverse prognostic effect of MALAT1 over-expression was obtained in different types of cancer (estrogen-dependent cancer: pooled HR = 2.656; urological cancer: pooled HR = 1.952; glioma: pooled HR = 2.315; digestive cancer: pooled HR = 2.451). The results of the study are consistent with most of the original studies, which suggest that MALAT1 may be associated with poor prognosis in malignant cancers [61,62,63]. However, the sensitivity analysis identified the studies of Droop et al. [36], which influenced the stability of the pooled results. The possible reasons were as follows. First, the heterogeneity across studies may be attributed to the difference of genetic background since the subjects of the study were from Germany. Second, Droop et al.’ the study involved two types of bladder cancer, including non-muscle-invasive tumours (NMIBC) and muscle-invasive bladder cancer (MIBC). NMIBC is essentially different from MIBC in tumor biology. Finally, the study is a small sample study. These potential confounders might account for the heterogeneity across studies. In addition, we need to explain that publication bias of LNM and differentiation may be due to small sample studies, which are susceptible to publication bias. Therefore, based on the above evidence, the abnormal regulation and prognostic utility of MALAT1 across multiple types of tumors suggests that MALAT may be a candidate biomarker for applying to therapeutic targets for clinical practice.
There are several limitations in the study. First, the cut-off values of high and low MALAT1 expression were different across studies. Second, the heterogeneity among studies may be due to different qRT-PCR primer sets. Third, significant heterogeneity may also be caused by confounding factors, such as cancer type, ethnicity, and other confounders. Fourth, several original studies did not provide complete data. Finally, the study might present “small-study effects” [91, 92]. For example, the last meta-analysis of HOXA11-AS demonstrated that small sample size studies with lacking of statistical power could obtain higher effect size compared with large sample studies [93]. Therefore, larger-scale studies are authorized to verify these results of the study.
Conclusions
In conclusion, the study revealed that over-expression MALAT1 might be an adverse biomarker for prognostic outcome, lymph node metastasis, distant metastasis, tumour size and TNM stage for cancer patients. MALAT1 might play a pivotal role in the tumorigenesis of multiple types of cancers. However, more high-quality larger-scale studies across ethnicities are warranted to explore the prognostic value and carcinogenic function of MALAT1 before it is applied to the treatment and management of cancer.
Abbreviations
- LncRNAs:
-
long noncoding RNAs
- MALAT1:
-
the metastasis-associated lung adenocarcinoma transcript 1
- qRT-PCR:
-
quantitative real-time polymerase chain reaction
- ISH:
-
in situ hybridization
- HWE:
-
hardy–weinberg equilibrium
- HR:
-
hazard ratio
- OS:
-
overall survival
- DFS:
-
disease-free survival
- RFS:
-
recurrence-free survival
- DSS:
-
disease-specific survival
- PFS:
-
progression-free survival
- GC:
-
gastric cancer
- GBC:
-
gallbladder cancer
- EC:
-
esophageal cancer
- PDAC:
-
pancreatic duct adenocarcinoma
- ESCC:
-
esophageal squamous cell carcinoma
- HCC:
-
hepatocellular carcinoma
- CRC:
-
colorectal cancer
- GBM:
-
glioblastoma multiforme
- EOC:
-
epithelial ovarian cancer
- BC:
-
breast cancer
- UC:
-
urothelial carcinoma
- ccRCC:
-
clear cell renal cell carcinoma
- SR:
-
serine/arginie-rich
- EMT:
-
epithelial–mesenchymal transition
- LNM:
-
lymph node metastasis
- ceRNA:
-
competitive endogenous RNA
References
Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59.
Hu X, Sood AK, Dang CV, Zhang L. The role of long noncoding RNAs in cancer: the dark matter matters. Curr Opin Genet Dev. 2017;48:8–15.
Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–87.
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–9.
Xu Y, Qiu M, Chen Y, Wang J, Xia W, Mao Q, Yang L, Li M, Jiang F, Xu L, et al. Long noncoding RNA, tissue differentiation-inducing nonprotein coding RNA is upregulated and promotes development of esophageal squamous cell carcinoma. Dis Esophagus. 2016;29(8):950–8.
Fu D, Zhang Y, Cui H. Long noncoding RNA CCAT2 is activated by E2F1 and exerts oncogenic properties by interacting with PTTG1 in pituitary adenomas. Am J Cancer Res. 2018;8(2):245–55.
Zhang M, Zhao Y, Zhang Y, Wang D, Gu S, Feng W, Peng W, Gong A, Xu M. LncRNA UCA1 promotes migration and invasion in pancreatic cancer cells via the Hippo pathway. Biochimica et biophysica acta. 2018;1864(5):1770–82.
Zhu Q, Luo Z, Lu G, Gui F, Wu J, Li F, Ni Y. LncRNA FABP5P3/miR-589-5p/ZMYND19 axis contributes to hepatocellular carcinoma cell proliferation, migration and invasion. Biochem Biophys Res Commun. 2018;498(3):551–8.
Ye H, Lin J, Yao X, Li Y, Lin X, Lu H. Overexpression of Long Non-Coding RNA NNT-AS1 Correlates with Tumor Progression and Poor Prognosis in Osteosarcoma. Cell Phys Biochem. 2018;45(5):1904–14.
Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat 1: its physiological and pathophysiological functions. RNA Biol. 2017;14(12):1705–14.
Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell reports. 2012;2(1):111–23.
Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–41.
Zuo Y, Li Y, Zhou Z, Ma M, Fu K. Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed Pharmacother Biomedecine pharmacotherapie. 2017;95:922–8.
Chen L, Yao H, Wang K, Liu X. Long non-coding RNA MALAT1 regulates ZEB1 expression by sponging miR-143-3p and promotes hepatocellular carcinoma progression. J Cell Biochem. 2017;118(12):4836–43.
Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9(3):e1003368.
Guo F, Guo L, Li Y, Zhou Q, Li Z. MALAT1 is an oncogenic long non-coding RNA associated with tumor invasion in non-small cell lung cancer regulated by DNA methylation. Int J Clin Exp Pathol. 2015;8(12):15903–10.
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20(6):1531–41.
Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol BioSyst. 2012;8(9):2289–94.
Zhang Y, Wang T, Huang HQ, Li W, Cheng XL, Yang J. Human MALAT-1 long non-coding RNA is overexpressed in cervical cancer metastasis and promotes cell proliferation, invasion and migration. J BUON. 2015;20(6):1497–503.
Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35(12):2731–9.
Wang F, Ren S, Chen R, Lu J, Shi X, Zhu Y, Zhang W, Jing T, Zhang C, Shen J, et al. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget. 2014;5(22):11091–102.
Sonohara F, Inokawa Y, Hayashi M, Yamada S, Sugimoto H, Fujii T, Kodera Y, Nomoto S. Prognostic value of long non-coding RNA HULC and MALAT1 Following the curative resection of hepatocellular carcinoma. Scientific Rep. 2017;7(1):16142.
Liu JH, Chen G, Dang YW, Li CJ, Luo DZ. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev. 2014;15(7):2971–7.
Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2015;36(4):2403–7.
Qiu G, Zhang XB, Zhang SQ, Liu PL, Wu W, Zhang JY, Dai SR. Dysregulation of MALAT1 and miR-619-5p as a prognostic indicator in advanced colorectal carcinoma. Oncol Lett. 2016;12(6):5036–42.
Yao W, Bai Y, Li Y, Guo L, Zeng P, Wang Y, Qi B, Liu S, Qin X, Li Y, et al. Upregulation of MALAT-1 and its association with survival rate and the effect on cell cycle and migration in patients with esophageal squamous cell carcinoma. Tumour Biol. 2016;37(4):4305–12.
Ma KX, Wang HJ, Li XR, Li T, Su G, Yang P, Wu JW. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour Biol. 2015;36(5):3355–9.
Shen J, Hodges TR, Song R, Gong Y, Calin GA, Heimberger AB, Zhao H. Serum HOTAIR and GAS5 levels as predictors of survival in patients with glioblastoma. Mol Carcinog. 2018;57(1):137–41.
Chen Q, Su Y, He X, Zhao W, Wu C, Zhang W, Si X, Dong B, Zhao L, Gao Y, et al. Plasma long non-coding RNA MALAT1 is associated with distant metastasis in patients with epithelial ovarian cancer. Oncol Lett. 2016;12(2):1361–6.
Meseure D, Vacher S, Lallemand F, Alsibai KD, Hatem R, Chemlali W, Nicolas A, De Koning L, Pasmant E, Callens C, et al. Prognostic value of a newly identified MALAT1 alternatively spliced transcript in breast cancer. Br J Cancer. 2016;114(12):1395–404.
Xu S, Sui S, Zhang J, Bai N, Shi Q, Zhang G, Gao S, You Z, Zhan C, Liu F, et al. Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int J Clin Exp Pathol. 2015;8(5):4881–91.
Li C, Cui Y, Liu LF, Ren WB, Li QQ, Zhou X, Li YL, Li Y, Bai XY, Zu XB. High expression of long noncoding RNA MALAT1 indicates a poor prognosis and promotes clinical progression and metastasis in bladder cancer. Clin Genitourin Cancer. 2017;15(5):570–6.
Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75(7):1322–31.
Zhang HM, Yang FQ, Chen SJ, Che J, Zheng JH. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2015;36(4):2947–55.
Handa H, Kuroda Y, Kimura K, Masuda Y, Hattori H, Alkebsi L, Matsumoto M, Kasamatsu T, Kobayashi N, Tahara KI, et al. Long non-coding RNA MALAT1 is an inducible stress response gene associated with extramedullary spread and poor prognosis of multiple myeloma. Br J Haematol. 2017;179(3):449–60.
Droop J, Szarvas T, Schulz WA, Niedworok C, Niegisch G, Scheckenbach K, Hoffmann MJ. Diagnostic and prognostic value of long noncoding RNAs as biomarkers in urothelial carcinoma. PLoS ONE. 2017;12(4):e0176287.
Cao X, Zhao R, Chen Q, Zhao Y, Zhang B, Zhang Y, Yu J, Han G, Cao W, Li J, et al. MALAT1 might be a predictive marker of poor prognosis in patients who underwent radical resection of middle thoracic esophageal squamous cell carcinoma. Cancer Biomarkers Sect A Dis Markers. 2015;15(6):717–23.
Duan W, Du L, Jiang X, Wang R, Yan S, Xie Y, Yan K, Wang Q, Wang L, Zhang X, et al. Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget. 2016;7(48):78850–8.
Jadaliha M, Zong X, Malakar P, Ray T, Singh DK, Freier SM, Jensen T, Prasanth SG, Karni R, Ray PS, et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget. 2016;7(26):40418–36.
Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6(12):1984–92.
Chen W, Xu XK, Li JL, Kong KK, Li H, Chen C, He J, Wang F, Li P, Ge XS, et al. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget. 2017;8(14):22783–99.
Gao KT, Lian D. Long non-coding RNA MALAT1 is an independent prognostic factor of osteosarcoma. Eur Rev Med Pharmacol Sci. 2016;20(17):3561–5.
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29(3):1810–6.
Li Y, Wu Z, Yuan J, Sun L, Lin L, Huang N, Bin J, Liao Y, Liao W. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett. 2017;395:31–44.
Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang Q, Ren M, Chen L, Yuan D, Zhang Y, et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget. 2016;7(35):56209–18.
Yang L, Bai HS, Deng Y, Fan L. High MALAT1 expression predicts a poor prognosis of cervical cancer and promotes cancer cell growth and invasion. Eur Rev Med Pharmacol Sci. 2015;19(17):3187–93.
Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P, Gu WL, Cai GX, Cai SJ. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7(6):3174–81.
Wang X, Sehgal L, Jain N, Khashab T, Mathur R, Samaniego F. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J Transl Med. 2016;14(1):346.
Huang NS, Chi YY, Xue JY, Liu MY, Huang S, Mo M, Zhou SL, Wu J. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer. Oncotarget. 2016;7(25):37957–65.
Huang C, Yu Z, Yang H, Lin Y. Increased MALAT1 expression predicts poor prognosis in esophageal cancer patients. Biomed Pharmacother Biomedecine pharmacotherapie. 2016;83:8–13.
Sun KK, Hu PP, Xu F. Prognostic significance of long non-coding RNA MALAT1 for predicting the recurrence and metastasis of gallbladder cancer and its effect on cell proliferation, migration, invasion, and apoptosis. J Cell Biochem. 2018;119(4):3099–110.
Cao S, Wang Y, Li J, Lv M, Niu H, Tian Y. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by suppressing miR-155 expression and activating FBXW7 function. Am J Cancer Res. 2016;6(11):2561–74.
Zhang R, Xia Y, Wang Z, Zheng J, Chen Y, Li X, Wang Y, Ming H. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;490(2):406–14.
Zhang Y, Chen Z, Li MJ, Guo HY, Jing NC. Long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 regulates the expression of Gli2 by miR-202 to strengthen gastric cancer progression. Biomed Pharmacother Biomedecine pharmacotherapie. 2017;85:264–71.
Jen J, Tang YA, Lu YH, Lin CC, Lai WW, Wang YC. Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol Cancer. 2017;16(1):104.
Wang Y, Zhang Y, Yang T, Zhao W, Wang N, Li P, Zeng X, Zhang W. Long non-coding RNA MALAT1 for promoting metastasis and proliferation by acting as a ceRNA of miR-144-3p in osteosarcoma cells. Oncotarget. 2017;8(35):59417–34.
Xiong Y, Wang J, Zhu H, Liu L, Jiang Y. Chronic oxymatrine treatment induces resistance and epithelialmesenchymal transition through targeting the long non-coding RNA MALAT1 in colorectal cancer cells. Oncol Rep. 2018;39(3):967–76.
Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res CR. 2015;34:7.
Li Q, Dai Y, Wang F, Hou S. Differentially expressed long non-coding RNAs and the prognostic potential in colorectal cancer. Neoplasma. 2016;63(6):977–83.
Jin C, Yan B, Lu Q, Lin Y, Ma L. The role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal carcinoma. Tumour Biol. 2016;37(3):4025–33.
Jin C, Yan B, Lu Q, Lin Y, Ma L. Reciprocal regulation of Hsa-miR-1 and long noncoding RNA MALAT1 promotes triple-negative breast cancer development. Tumour Biol. 2016;37(6):7383–94.
Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36(3):1477–86.
Luo F, Sun B, Li H, Xu Y, Liu Y, Liu X, Lu L, Li J, Wang Q, Wei S, et al. A MALAT1/HIF-2alpha feedback loop contributes to arsenite carcinogenesis. Oncotarget. 2016;7(5):5769–87.
Li J, Gao J, Tian W, Li Y, Zhang J. Long non-coding RNA MALAT1 drives gastric cancer progression by regulating HMGB2 modulating the miR-1297. Cancer Cell Int. 2017;17:44.
Wang SH, Zhang WJ, Wu XC, Zhang MD, Weng MZ, Zhou D, Wang JD, Quan ZW. Long non-coding RNA Malat1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206. Oncotarget. 2016;7(25):37857–67.
Wu L, Wang X, Guo Y. Long non-coding RNA MALAT1 is upregulated and involved in cell proliferation, migration and apoptosis in ovarian cancer. Exp Ther Med. 2017;13(6):3055–60.
Miao Y, Fan R, Chen L, Qian H. Clinical significance of long non-coding RNA MALAT1 expression in tissue and serum of breast cancer. Ann Clin Lab Sci. 2016;46(4):418–24.
Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, Guo Z, Wang L. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep. 2014;32(6):2485–92.
Chou J, Wang B, Zheng T, Li X, Zheng L, Hu J, Zhang Y, Xing Y, Xi T. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem Biophys Res Commun. 2016;472(1):262–9.
Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding Q, Weng H, Shu YJ, Liu TY, Jiang L, et al. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther. 2014;15(6):806–14.
Jin Y, Feng SJ, Qiu S, Shao N, Zheng JH. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med Pharmacol Sci. 2017;21(14):3176–84.
Liu P, Yang H, Zhang J, Peng X, Lu Z, Tong W, Chen J. The lncRNA MALAT1 acts as a competing endogenous RNA to regulate KRAS expression by sponging miR-217 in pancreatic ductal adenocarcinoma. Scientific Rep. 2017;7(1):5186.
Ren S, Liu Y, Xu W, Sun Y, Lu J, Wang F, Wei M, Shen J, Hou J, Gao X, et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol. 2013;190(6):2278–87.
Wang D, Ding L, Wang L, Zhao Y, Sun Z, Karnes RJ, Zhang J, Huang H. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget. 2015;6(38):41045–55.
Deng QJ, Xie LQ, Li H. Overexpressed MALAT1 promotes invasion and metastasis of gastric cancer cells via increasing EGFL7 expression. Life Sci. 2016;157:38–44.
Wang W, Zhu Y, Li S, Chen X, Jiang G, Shen Z, Qiao Y, Wang L, Zheng P, Zhang Y. Long noncoding RNA MALAT1 promotes malignant development of esophageal squamous cell carcinoma by targeting beta-catenin via Ezh2. Oncotarget. 2016;7(18):25668–82.
Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, Zhou L, Zhou C, Yuan Q, Yang M. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290(7):3925–35.
Guo F, Jiao F, Song Z, Li S, Liu B, Yang H, Zhou Q, Li Z. Regulation of MALAT1 expression by TDP43 controls the migration and invasion of non-small cell lung cancer cells in vitro. Biochem Biophys Res Commun. 2015;465(2):293–8.
Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, Sui H, Tang Y, Wang Y, Liu N, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111(4):736–48.
Sun R, Qin C, Jiang B, Fang S, Pan X, Peng L, Liu Z, Li W, Li Y, Li G. Down-regulation of MALAT1 inhibits cervical cancer cell invasion and metastasis by inhibition of epithelial–mesenchymal transition. Mol BioSyst. 2016;12(3):952–62.
Li J, Wang J, Chen Y, Li S, Jin M, Wang H, Chen Z, Yu W. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR-204. Am J Cancer Res. 2016;6(5):1099.
Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, Li ZG. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochem Biophys Acta. 2015;1852(1):166–74.
Li L, Chen H, Gao Y, Wang YW, Zhang GQ, Pan SH, Ji L, Kong R, Wang G, Jia YH, et al. Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol Cancer Ther. 2016;15(9):2232–43.
Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S. MALAT1 long non-coding RNA in cancer. Biochem Biophys Acta. 2016;1859(1):192–9.
Amodio N, Raimondi L, Juli G, Stamato MA, Caracciolo D, Tagliaferri P, Tassone P. MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J Hematol Oncol. 2018;11(1):63.
Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–38.
Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147(4):773–88.
Liu J, Peng WX, Mo YY, Luo D. MALAT1-mediated tumorigenesis. Front Biosci (Landmark edition). 2017;22:66–80.
Shuai P, Zhou Y, Gong B, Jiang Z, Yang C, Yang H, Zhang D, Zhu S. Long noncoding RNA MALAT1 can serve as a valuable biomarker for prognosis and lymph node metastasis in various cancers: a meta-analysis. SpringerPlus. 2016;5(1):1721.
Lin Y, Fang Z, Lin Z, Li Z, Zhao J, Luo Y, Xu B. The prognostic impact of long noncoding RNA HOTAIR in leukemia and lymphoma: a meta-analysis. Hematology. 2018;7:1–8.
Serghiou S, Kyriakopoulou A, Ioannidis JP. Long noncoding RNAs as novel predictors of survival in human cancer: a systematic review and meta-analysis. Mol Cancer. 2016;15(1):50.
Ioannidis JP, Panagiotou OA. Comparison of effect sizes associated with biomarkers reported in highly cited individual articles and in subsequent meta-analyses. JAMA. 2011;305(21):2200–10.
Mu S, Ai L, Fan F, Sun C, Hu Y. Prognostic and clinicopathological significance of long noncoding RNA HOXA11-AS expression in human solid tumors: a meta-analysis. Cancer Cell Int. 2018;18:1.
Authors’ contributions
JL, ZHY and HL conceived and wrote the paper; JL, XTL and MG collected and analyzed the data; JL, ZGC, HL, MG, ZTY and YHB revised the whole paper. All authors have reviewed the final version of the manuscript and approved to submit to your journal. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All the original data presented in additional files. Please contact authors for other data request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This study was supported by National Natural Science Foundation of China (No. 81673261).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Table S1.
The characteristics of the included studies in the meta-analysis.
Additional file 2: Table S2.
Characteristics of studies for survival outcomes included in the meta-analysis.
Additional file 3: Table S3.
Association between MALAT1 expression and clinicopathological features of cancers.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, J., Cui, Z., Li, H. et al. Clinicopathological and prognostic significance of long noncoding RNA MALAT1 in human cancers: a review and meta-analysis. Cancer Cell Int 18, 109 (2018). https://doi.org/10.1186/s12935-018-0606-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-018-0606-z