Abstract
Background
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), a newly discovered long intergenic noncoding RNA (lincRNA), has been reported to be aberrantly expressed in various cancers, and may serve as a novel potential biomarker for cancer prognosis. This meta-analysis was conducted to investigate the effects of MALAT1 on cancer prognosis and lymph node metastasis.
Methods
A quantitative meta-analysis was performed using a systematic search of PubMed, Medline and Web of Science databases to identify eligible papers on prognostic value of MALAT1 in cancers. The pooled hazard ratios (HRs) or odds ratios (OR) with a 95 % confidence interval (95 % Cl) were calculated to evaluate its prognostic value.
Results
A total of 2094 patients from 17 studies between 2003 and 2016 were included. The results revealed that elevated MALAT1 expression was significantly associated with poor overall survival (OS) in 11 types of cancers (HR = 1.91, 95 % CI 1.49–2.34). Furthermore, subgroup analysis indicated that region of study (Germany, Japan or China), cancer type (digestive system cancers, urinary system cancers or respiratory system cancers) and sample size (more or less than 100) did not alter the significant predictive value of MALAT1 in OS from various types of cancer. In addition, upregulation of MALAT1 expression was significantly associated with poor disease-free survival (HR = 2.29, 95 % CI 1.24–3.35), and recurrence-free survival (HR = 2.09, 95 % CI 0.81–3.37). The results showed that the incidence of lymph node metastasis was higher in high MALAT1 expression group than that in low MALAT1 expression group (OR = 1.67, 95 % CI 1.30–2.15).
Conclusions
This meta-analysis revealed that elevated MALAT1 expression may serve as a novel predictive biomarker for poor survival and lymph node metastasis in different types of cancer.
Similar content being viewed by others
Background
Cancer has now become a major cause of morbidity and mortality in most regions worldwide, and the 5-year survival rate remains low in a wide types of cancer (Bray et al. 2013). With the development of genome-wide sequencing technology, numerous investigators are searching for biomarkers that may help with the clinical diagnosis or prognostic prediction of various cancers. Recently, long noncoding RNAs (lncRNAs) have caught increasing attention, and might act as potential valuable biomarkers for cancer diagnosis or potential targets for cancer therapy (Esteller 2011; Gibb et al. 2011).
LncRNAs are generally defined as transcribed RNA molecules with a length greater than 200 nt, and most lack protein coding capability. However, accumulating evidences have suggested that lncRNAs participated in a wide range of biological pathways. LncRNAs could gene expression through diverse molecular mechanisms, including transcriptional and post–transcriptional processing, chromatin modification and epigenetics, genomic imprinting, protein activity modulation and protein localization. Recent studies found that dysregulation of lncRNAs is always associated with cancer development and progression (Zhang et al. 2014; Wang et al. 2014). Importantly, lncRNAs could play important regulators in various cancer-related processes such as modulation of apoptosis and proliferation, drug resistance and the process of epithelial-mesenchymal transition (EMT) (Yang et al. 2014). However, the role of most lncRNAs in cancer progression and prognosis still remains unclear.
Metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) is a highly conserved nuclear-abundant lncRNA with more than 8000 nucleotides derived from chromosome 11q13. It was the first lncRNA that identified as a prognostic biomarker for early stage non-small cell lung cancer (Ji et al. 2003; Guru et al. 1997; van Asseldonk et al. 2000). Up-regulation of MALAT1 expression was reported in a variety of cancers, including non-small cell lung cancer (Ji et al. 2003; Schmidt et al. 2011), hepatocellular carcinoma (Lai et al. 2012), gastric cancer (Okugawa et al. 2014), pancreatic cancer (Liu et al. 2014; Pang et al. 2015), colorectal cancer (Zheng et al. 2014), clear cell renal cell carcinoma (Zhang et al. 2015), esophageal cancer (Hu et al. 2015; Huang et al. 2016a), gallbladder cancer (Wang et al. 2016), bladder cancer (Fan et al. 2014), osteosarcoma (Dong et al. 2015) and breast cancer (Huang et al. 2016b; Miao et al. 2016). MALAT1 is thought to regulate tumor cell viability, invasiveness and migration (Yoshimoto et al. 2016). Recent study found that knock-down of MALAT1 expression could inhibit pancreatic cancer cell proliferation, migration, invasion and the process of epithelial-mesenchymal transition, and induce cell apoptosis and G2/M cell cycle arrest in vitro (Jiao et al. 2014). Moreover, Dong et al. found that that MALAT1 might promote the proliferation and metastasis of osteosarcoma cells by activating the PI3 K/Akt signaling pathway (Dong et al. 2015). Lai et al. demonstrated that downregulation of MALAT1 expression in HepG2 cells suppressed cell viability, motility, and invasiveness and elevated the sensitivity to apoptosis (Lai et al. 2012).
A number of studies have revealed that overexpression of MALAT1 was significantly associated with high-risk grade, metastasis and patients’ survival in many types of cancers (Xia et al. 2016; Wang et al. 2016; Li et al. 2016). Although MALAT1 expression is considered to relating to clinical prognosis of multiple cancers, the specific impact of MALAT1 expression on the outcome of cancer patients still needs to be further investigated. Therefore, we conducted a systematic review and quantitative meta-analysis to further investigate the prognostic role of MALAT1 overexpression in human cancers.
Methods
Literature collection
The selected literatures were determined via an electronic search of PubMed, EMBASE, Web of Science, Ovid and Cochrane library databases using these following terms: “MALAT1”, “NEAT2”, “long intergenic noncoding RNA”, “lincRNA”, “lncRNA”, “noncoding RNA”, “cancer”, ‘‘carcinoma”, “neoplasm”, “outcome”, “prognosis”, “prognostic”, “mortality”, “survival”, “metastasis”, “recurrence” and “lymph node metastasis”. The literature search, which was performed by two independent researchers, was limited to the English language. The last search was updated in August 28, 2016. The reference lists of the retrieved articles were searched manually.
Inclusion and exclusion criteria
The studies were considered eligible if they met the following criteria: any type of human cancer was studied; studies investigating the prognostic role of MALAT1 in various cancers; the levels of MALAT1 expression in cancerous tissues were detected; patients were grouped according to the levels of MALAT1 expression; a link between MALAT1 expression and clinicopathologic parameters was included; studies providing sufficient data to estimate the HRs with corresponding 95 % CI for overall survival (OS), disease specific survival (DSS), disease-free survival (DFS) or disease-free survival (RFS); and studies published in English. Exclusion criteria were as follows: letters, editorials, expert opinions, case reports and reviews; studies only investigated the molecular structure and functions of MALAT1; studies without usable data for further analysis; and duplicate publications.
Data extraction
Two investigators (PS, YZ) independently evaluated and extracted data from each identified studies based on criteria of inclusion and exclusion. Corresponding authors were contacted to clarify missing or ambiguous data. The following information was carefully extracted: first author, publication date, country, ethnicity, tumor type, sample size, detection method, follow-up period, cut-off value, the number of high and low MALAT1 expression, the number of patients with lymph node metastasis, survival analysis methodology, HRs with corresponding 95 % CIs for OS, DSS, RFS or DFS. HRs with corresponding 95 % CIs were preferentially extracted from univariate or multivariate analyses. If these were not available, the HR estimates were calculated from Kaplan–Meier survival curves using Engauge Digitizer V4.1 (http://sourceforge.net).
Statistical analyses
All data analyses were performed with STATA statistical software version 14.0 (Stata Corporation, College Station, TX, USA). Pooled HRs with 95 % CIs were used to estimate the strength of the link between MALAT1 and clinical prognosis. X 2-based Cochran Q test and Higgins I 2 statistic were utilized to determine the heterogeneity among the included studies. P value <0.05 in combination with I 2 value >50 % was considered significant heterogeneity. Random-effects models were applied in cases with significant heterogeneity. Subgroup analysis and sensitivity analysis were performed to dissect the heterogeneity. In addition, publication bias was determined using Egger’s linear regression test and Begg’s funnel plots analysis. P-value <0.05 was considered statistically significant.
Results
Studies characteristics
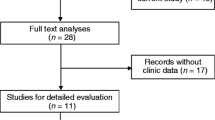
A total of 255 relevant articles were identified based on electronic databases including PubMed, EMBASE, Web of Science, Ovid and Cochrane library. On the basis of the inclusion criteria, 17 eligible papers were enrolled in this meta-analysis (Fig. 1). The characteristics of the included studies are summarized in Table 1. A total of 2094 patients from 17 studies between 2003 and 2016 were included. The regions represented in the studies including Germany (n = 2), Japan (n = 2) and China (n = 13). Among these studies, the sample size ranged between 19 and 352, and 10 studies enrolled more than 100. Eleven types of human cancer were recorded including non-small cell lung cancer (NSCLC), hepatocellular carcinoma (HCC), gastric cancer (GC), pancreatic ductal adenocarcinoma (PDAC), colorectal cancer (CRC), clear cell renal cell carcinoma (ccRCC), esophageal squamous cell carcinoma (ESCC), esophageal cancer (EC), gallbladder cancer (GBC), bladder cancer, osteosarcoma and breast cancer. The levels of MALAT1 expression were detected by reverse transcription quantitative polymerase chain reaction (qRT-PCR) or situ hybridization (ISH). The cut-off values in the included studies were inconsistent partly due to different detection methods. The clinical outcomes were also recoded including 12 studies for OS, 2 for DFS, 2 for PFS, 1 for DSS, and 12 for lymph node metastasis. HRs with the corresponding 95 % CIs were extracted from the original data including 4 studies for OS, 3 for DFS, 1 for DFS, 1 for RFS, and 1 for DFS, and calculated from Kaplan–Meier Curves in 8 studies for OS, 1 for DFS and 1 for RFS.
Association between MALAT1 expression and survival in different types of cancers
A total of 12 studies including 1549 patients were recruited to assess the effect of MALAT1 overexpression on OS in different types of cancer. It was suggested that elevated MALAT1 expression predicted a poor outcome for OS in 12 types of cancers (pooled HR = 1.91, 95 % CI 1.49–2.34) with no heterogeneity (I2 = 0.0 %, P = 0.728) (Fig. 2). Furthermore, subgroup analysis was also conducted based on region of study, cancer type and sample size. Stratified analyses by region of study indicated that the different effect of elevated MALAT1 on OS among different countries, including Germany (pooled HR = 1.72, 95 % CI 0.86–2.57), Japan (pooled HR = 1.55, 95 % CI 0.72–2.38), and China (pooled HR = 2.22, 95 % CI 1.60–2.84) (Fig. 2a). A significant relationship was observed between MALAT1 overexpression and poor OS in the studies for digestive system cancers (pooled HR = 1.77, 95 % CI 1.18–2.35), urinary system cancers (pooled HR = 2.95, 95 % CI 1.43–4.46), and respiratory system cancers (pooled HR = 1.72, 95 % CI 0.86–2.57) (Fig. 2b). In addition, the negative effect of MALAT1 overexpression on predicting poor OS was shown in studies including <100 patients (pooled HR = 2.89, 95 % CI 1.05–4.73) as well as those with >100 patients (pooled HR = 1.86, 95 % CI 1.42–2.30) (Fig. 2c).
In addition, two studies including 224 patients reported HRs for DFS (Fig. 3b). High level of MALAT1 expression was observed to be a negative factor for DFS (pooled HR = 2.29, 95 % CI 1.24–3.35) with no heterogeneity (I2 = 0.0 %, P = 0.375). Two studies including 368 patients reported HRs for RFS. Upregulation of MALAT1 expression was associated with poor RFS (pooled HR = 2.09, 95 % CI 0.81–3.37) with no heterogeneity (I2 = 0.0 %, P = 0.346).
Association between MALAT1 expression and lymph node metastasis
Twelve studies comprising 798 patients were pooled for the meta-analysis of lymph node metastasis. In high MALAT1 expression group, 393 patients (49.2 %) with cancer had lymph node metastasis, while only 405 patients (50.8 %) in low MALAT1 expression group. Due to the presence of heterogeneity (I2 = 73.1 %, P < 0.001), we compared the incidence of lymph node metastasis between these two groups with the random-effects model. The results found that the incidence of lymph node metastasis was higher in high MALAT1 expression group than that in low MALAT1 expression group (pooled OR = 1.67, 95 % CI 1.30–2.15) (Fig. 3a). This result demonstrated that the patients with high levels of MALAT1 expression are more prone to lymph node metastasis in different systemic cancers.
Publication bias and sensitivity analysis
To evaluate publication bias in this meta-analysis, the included studies were conducted using Egger’s linear regression test and Begg’s funnel plots analysis. The results of Egger’s linear regression test revealed no publication bias for the analysis of OS (t = 2.10, P = 0.062). The plots were also asymmetrically inverted funnels (Fig. 4). Due to the small number of studies, publication bias was not analyzed in the DFS and RFS groups. In addition, sensitivity analysis indicated that the pooled HR for deregulated MALAT1 associated with OS was not significantly affected by the exclusion of any of the studies (data not shown).
Discussion
Although these long noncoding transcripts were once considered to be simply transcriptional noise or cloning artifacts, numerous studies have suggested that lncRNAs are emerging as new players in diverse human diseases, especially cancers (Young and Ponting 2013; Qi and Du 2013). Many studies have demonstrated that lncRNAs are involved in various biological processes, including modulation of apoptosis and proliferation, drug resistance, the process of EMT, cancer progression and metastasis (Hajjari et al. 2014; Yang et al. 2014). An increasing number of studies have focused on the involvement of lncRNAs in cancer development and progression, and proposed them as potential biomarkers for cancer diagnosis or potential targets for cancer therapy (Cui et al. 2016; Yuan et al. 2016; Xie et al. 2016; Liz and Esteller 2016).
MALAT1, regarded as one of the most familiar oncogenic lncRNAs, was overexpressed in various cancers (Yoshimoto et al. 2016). MALAT1 could promote tumor progression through multiple mechanisms in various types of cancer. Increasing reports have demonstrated that MALAT1 could function as a promoter of cell proliferation and metastasis (Thum and Fiedler 2014). Jiao et al. reported that MALAT1 could possibly inhibit G2/M cell cycle arrest, thus promoting EMT in pancreatic cancer (Jiao et al. 2014). Then, Yao et al. found that MALAT1 knockdown induced a decrease in proliferation-enhanced apoptosis, inhibited invasion, and reduced colony formation and led to cell cycle arrest at the G2/M phase in esophageal squamous cell carcinoma (Yao et al. 2016). Xu et al. (2016) revealed that MALAT1 promotes the proliferation of chondrosarcoma cell by activating Notch-1 pathway. Besides, Ji et al. 2013 demonstrated that resveratrol could inhibit invasion and metastasis of colorectal cancer cells though MALAT1 mediated Wnt/beta-catenin pathway . However, the mechanisms underlying aberrant expression of MALAT1 in cancers remain elusive. Some researcher discovered that upregulation of MALAT1 was mediated by the transcription factor Sp1 in A549 lung cancer cells (Li et al. 2015). Additionally, Wang et al. found that silencing of MALAT1 by miR-101 and miR-217 inhibited proliferation, migration, and invasion of esophageal cacner cells (Wang et al. 2015).
Recent studies found that overexpression of MALAT1 correlated with poor prognosis of patients with nearly all types of cancer (Zheng et al. 2014; Zhang et al. 2015; Wang et al. 2016). However, the perplexity and inconsistence arise from a wide range of studies due to heterogeneity. Therefore, a meta-analysis of 2094 cancer patients from 17 studies was undertaken to assess the prognostic value of MALAT1 expression in this study. The pooled data revealed that increased MALAT1 expression was significantly associated with poor OS, DFS and RFS, indicating that MALAT1 has been shown to contribute to cancer progression and considered as a promising prog-nostic biomarker for cancer patients. Furthermore, subgroup analysis indicated that region of study (Germany, Japan or China), cancer type (digestive system cancers, urinary system cancers or respiratory system cancers) and sample size (more or less than 100) did not alter the significant predictive value of MALAT1 in OS from various types of cancer. In addition, MALAT1 was originally discovered to be a marker for metastasis development in early stage lung adenocarcinoma (Ji et al. 2003), and more recently it was shown to be a metastasis-related marker both in squamous cell carcinoma of the lung and hepatocellular carcinoma (Schmidt et al. 2011; Lai et al. 2012). In this present study, we further verified that overexpression of MALAT1 is associated with the incidence of lymph node metastasis in many types of cancer.
Some limitations of this meta-analysis should be acknowledged due to the discrete data across these clinical studies. The major limitation of this meta-analysis was that we only included studies reporting HR or survival curves (except the data that cannot be calculated), and consequently some publications reporting on the prognostic value of MALAT1 were excluded, thus the selection bias might be appeared. Secondly, a part of studies, especially in subgroups analyses, was lightly relative. Thirdly, only summarized data rather than individual patient data were used. Fourthly, some of the HRs could not be directly obtained from the primary studies, we reconstruct the survival curves to extract the HR estimates or calculate them using the reported data. Finally, the cut-off value of high and low MALAT1 expression varied in different studies. Therefore, it is possible that our results might overestimate the prognostic effects of abnormal MALAT1 expression on survival and lymph node metastasis in different types of cancer. We believed that more clinical studies should be conducted to evaluate potential prognostic role of MALAT1 in other types of cancer that have not been included.
Conclusion
This meta-analysis explored that elevated MALAT1 expression is common to various types of cancer and might act as a novel predictive factor of poor prognosis and lymph node metastasis in different type of cancer. Furthermore, the functional role of MALAT1 in the regulation of cell proliferation, apoptosis and metastasis suggested that MALAT1 may play a key role in tumor progression. Nevertheless, it is still necessary to conduct larger-size and better design studies to confirm our results.
References
Bray F, Ren JS, Masuyer E, Ferlay J (2013) Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer 132(5):1133–1145
Cui M, You L, Ren X, Zhao W, Liao Q, Zhao Y (2016) Long non-coding RNA PVT1 and cancer. Biochem Biophys Res Commun 471(1):10–14
Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X (2015) MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3 K/Akt pathway. Tumour Biol 36(3):1477–1486
Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12(12):861–874
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y (2014) TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res 20(6):1531–1541
Gibb EA, Brown CJ, Lam WL (2011) The functional role of long non-coding RNA in human carcinomas. Mol Cancer 10:38
Guru SC, Agarwal SK, Manickam P, Olufemi SE, Crabtree JS, Weisemann JM, Kester MB, Kim YS, Wang Y, Emmert-Buck MR, Liotta LA, Spiegel AM, Boguski MS, Roe BA, Collins FS, Marx SJ, Burns L, Chandrasekharappa SC (1997) A transcript map for the 2.8-Mb region containing the multiple endocrine neoplasia type 1 locus. Genome Res 7(7):725–735
Hajjari M, Khoshnevisan A, Shin YK (2014) Molecular function and regulation of long non-coding RNAs: paradigms with potential roles in cancer. Tumour Biol 35(11):10645–10663
Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K (2015) Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res 34:7
Huang C, Yu Z, Yang H, Lin Y (2016a) Increased MALAT1 expression predicts poor prognosis in esophageal cancer patients. Biomed Pharmacother 83:8–13
Huang NS, Chi YY, Xue JY, Liu MY, Huang S, Mo M, Zhou SL, Wu J (2016b) Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer. Oncotarget 7(25):37957–37965
Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C (2003) MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22(39):8031–8041
Ji Q, Liu X, Fu X, Zhang L, Sui H, Zhou L, Sun J, Cai J, Qin J, Ren J, Li Q (2013) Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/beta-catenin signal pathway. PLoS ONE 8(11):e78700
Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, Guo Z, Wang L (2014) Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep 32(6):2485–2492
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS (2012) Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol 29(3):1810–1816
Li S, Wang Q, Qiang Q, Shan H, Shi M, Chen B, Zhao S, Yuan L (2015) Sp1-mediated transcriptional regulation of MALAT1 plays a critical role in tumor. J Cancer Res Clin Oncol 141(11):1909–1920
Li T, Mo X, Fu L, Xiao B, Guo J (2016) Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget 7(8):8601–8612
Liu JH, Chen G, Dang YW, Li CJ, Luo DZ (2014) Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev 15(7):2971–2977
Liz J, Esteller M (2016) lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta 1859(1):169–176
Miao Y, Fan R, Chen L, Qian H (2016) Clinical significance of long non-coding RNA MALAT1 expression in tissue and serum of breast cancer. Ann Clin Lab Sci 46(4):418–424
Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, Goel A (2014) Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis 35(12):2731–2739
Pang EJ, Yang R, Fu XB, Liu YF (2015) Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol 36(4):2403–2407
Qi P, Du X (2013) The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol 26(2):155–165. doi:10.1038/modpathol.2012.160
Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L, Wiebe K, Berdel WE, Wiewrodt R, Muller-Tidow C (2011) The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol 6(12):1984–1992
Thum T, Fiedler J (2014) LINCing MALAT1 and angiogenesis. Circ Res 114(9):1366–1368
van Asseldonk M, Schepens M, de Bruijn D, Janssen B, Merkx G, Geurts van Kessel A (2000) Construction of a 350-kb sequence-ready 11q13 cosmid contig encompassing the markers D11S4933 and D11S546: mapping of 11 genes and 3 tumor-associated translocation breakpoints. Genomics 66(1):35–42
Wang L, He JH, Han ZP (2014) Characteristics of PVT1 and its roles in diseases. Chin Med Sci J 29(4):236–238
Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, Zhou L, Zhou C, Yuan Q, Yang M (2015) Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem 290(7):3925–3935
Wang SH, Zhang WJ, Wu XC, Zhang MD, Weng MZ, Zhou D, Wang JD, Quan ZW (2016) Long non-coding RNA Malat1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206. Oncotarget 7(25):37857–37867
Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang Q, Ren M, Chen L, Yuan D, Zhang Y, Liu M, Gong Q, Bi F (2016) The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget 7(35):56209–56218
Xie Z, Chen X, Li J, Guo Y, Li H, Pan X, Jiang J, Liu H, Wu B (2016) Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer. Oncotarget 7(18):25408–25419
Xu F, Zhang ZQ, Fang YC, Li XL, Sun Y, Xiong CZ, Yan LQ, Wang Q (2016) Metastasis-associated lung adenocarcinoma transcript 1 promotes the proliferation of chondrosarcoma cell via activating Notch-1 signaling pathway. Onco Targets Ther 9:2143–2151
Yang G, Lu X, Yuan L (2014) LncRNA: a link between RNA and cancer. Biochim Biophys Acta 1839 11:1097–1109
Yao W, Bai Y, Li Y, Guo L, Zeng P, Wang Y, Qi B, Liu S, Qin X, Li Y, Zhao B (2016) Upregulation of MALAT-1 and its association with survival rate and the effect on cell cycle and migration in patients with esophageal squamous cell carcinoma. Tumour Biol 37(4):4305–4312
Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S (2016) MALAT1 long non-coding RNA in cancer. Biochim Biophys Acta 1:192–199
Young RS, Ponting CP (2013) Identification and function of long non-coding RNAs. Essays Biochem 54:113–126
Yuan J, Yue H, Zhang M, Luo J, Liu L, Wu W, Xiao T, Chen X, Chen X, Zhang D, Xing R, Tong X, Wu N, Zhao J, Lu Y, Guo M, Chen R (2016) Transcriptional profiling analysis and functional prediction of long noncoding RNAs in cancer. Oncotarget 7(7):8131–8142
Zhang A, Xu M, Mo YY (2014) Role of the lncRNA-p53 regulatory network in cancer. J Mol Cell Biol 6(3):181–191
Zhang HM, Yang FQ, Chen SJ, Che J, Zheng JH (2015) Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol 36(4):2947–2955
Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P, Gu WL, Cai GX, Cai SJ (2014) High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol 7(6):3174–3181
Authors’ contributions
YZ carried out this meta-analysis, collected the data, analyzed and interpreted the data as well as participated in the sequence alignment to articulate the manuscript. PS assessed all the eligible studies, extracted the data and performed statistical analyses. SKZ designed and drafted the article, made critical revision for important intellectual content with thorough supervision. DDZ aligned the article sequentially, designed, coordinated and helped to draft the manuscript. Other authors performed statistical analyses, and aided in finalizing the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This study was supported by grants from the Nation Natural Science Foundation of China (No. 81271007). The authors alone are responsible for the content and the writing of the paper.
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Ping Shuai and Yu Zhou contributed equally to this work
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Shuai, P., Zhou, Y., Gong, B. et al. Long noncoding RNA MALAT1 can serve as a valuable biomarker for prognosis and lymph node metastasis in various cancers: a meta-analysis. SpringerPlus 5, 1721 (2016). https://doi.org/10.1186/s40064-016-3342-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40064-016-3342-7