Abstract
Background
Neuroblastoma (NB) is the most common malignant tumor originating from the extracranial sympathetic nervous system in children. The molecular mechanisms underlying this disease are complex, and not completely understood.
Methods
Quantitative real-time PCR (qRT-PCR) was applied to quantify the expression of miR-20a-5p and its target gene ATG7 in clinical NB tissues. The biological function of miR-20a-5p and ATG7 in SH-SY5Y cells was investigated through in vitro studies (Real-Time cell kinetic analyzer, colony formation assay, caspase-Glo 3/7 assay and western blotting). The luciferase reporter assay was conducted to verify the biological relationship between miR-20a-5p and ATG7.
Results
Here we found that miR-20a-5p expression was significantly downregulated whereas its target autophagy-related gene 7 (ATG7) was increased along with clinical staging of NB progression. Correlation analysis showed that miR-20a-5p had a negative correlation trend with ATG7. In SH-SY5Y cells, forced expression of miR-20a-5p suppressed ATG7 expression, autophagy initiation and cellular proliferation while promoted apoptosis, suggesting a potential association between miR-20a-5p and ATG7. Further bioinformatic target prediction combined with protein expression and luciferase reporter assay verified that miR-20a-5p inhibited ATG7 by directly binding to its 3′-UTR, confirming the involvement of miR-20a-5p in the regulation of ATG7 in NB.
Conclusions
These results clarified that miR-20a-5p inhibited cell proliferation and promoted apoptosis through negative regulation of ATG7 and thus autophagy suppression in SH-SY5Y cells. Therefore, defining the context-specific roles of autophagy in NB and regulatory mechanisms involved will be critical for developing autophagy-targeted therapeutics against NB. Both miR-20a-5p and ATG7 would be potential therapeutic targets for future NB treatment.
Similar content being viewed by others
Background
Neuroblastoma (NB) is the most common extracranial solid tumour occurring in children [1], which accounts for more than 15% of all pediatric oncology deaths. Pediatric patients with NB have a poor prognosis despite receiving multimodal treatments such as surgery, radiotherapy, photodynamic therapy, and chemotherapy. Half of neuroblastoma cases are classified as high-risk for disease relapse, with long-term survival less than 40% [2]. Even when whole-genome sequencing of neuroblastoma was conducted, few recurrent gene alterations (MYCN, ALK, ATRX and TERT) were identified [3,4,5]. The unclear pathogenesis of neuroblastoma impedes the development of therapeutic drugs discovery and effective tumor therapy. With decades of efforts, only Unituxin (dinutuximab) was approved by the US Food and Drug Administration as a novel targeted drug in 2015, which can prolong the survival of patients with high-risk NB [6]. Therefore, clear clarification of mechanisms underlying NB progression is urgently needed.
MicroRNAs (miRNAs) are small non-coding RNAs of 19–25 nucleotides in length, serving as post-transcriptional regulators of gene expression [7]. Functionally, miRNAs can regulate genes involved in diverse biological processes, such as cell proliferation, development, differentiation and apoptosis [8]. Pathologically, abnormal microRNA expression is involved in tumorigenesis [9]. In NB, miR-23a, miR-421 and miR-558 promoted tumor growth, invasion, metastasis and induced angiogenisis [10,11,12]. Recently, miR-451 was reported reduced in NB tissues and correlated with tumour size, lymph node metastasis, tumour-node-metastasis (TNM) stage and distant metastases [13]. These studies indicated that miRNAs contributed to diverse processes in NB, acting as oncogenes and/or tumor suppressors. The miR-20a-5p is a 23-nucleotides-length non-coding RNA. Functionally, various studies have been conducted to investigate the effects of miR-20a-5p in types of tumors. Some of the results demonstrated that miR-20a-5p promoted radio-resistance in nasopharyngeal cancer cells [14], promoted colorectal cancer invasion and metastasis [15], and repressed multi-drug resistance in osteosarcoma [16]. However, the accurate expression, function and mechanism in tumor, especially in pediatric NB, are largely unclear.
Accumulating reports have revealed that miRNAs can modulate autophagic pathways [17]. Autophagy is an intracellular process highly regulated by autophagy-related genes (ATGs) for lysosomal degradation and recycling of proteins and organelles [18]. Autophagy dysfunction can lead to severe pathological states, such as neurodegenerative diseases and particularly cancer [19, 20]. As a pivotal regulator in autophagy initiation and autophagosome formation, ATG7 contributes to tumor cell proliferation, cell death and drug resistance [21]. Recently, miR-375 was reported to inhibit autophagy and reduce viability via ATG7 in hepatocellular carcinoma cells under hypoxic conditions [22]. Moreover, miR-200b could downregulate ATG12, suppress autophagy and enhance chemosensitivity both in vivo and in vitro [23]. In pediatric NB, autophagy was also associated with chemoresistance and proliferation [24, 25], but the involved underlying mechanisms were not clear. Although evidence has linked miRNAs to autophagy, it is far from clear whether miR-20a-5p contributes to the regulatory network of autophagy in NB.
To investigate the function and underlying mechanism of miR-20a-5p in NB proliferation, this study was then conducted. We found that miR-20a-5p was significantly downregulated, while ATG7 was upregulated along with clinical staging of NB progression. MiR-20a-5p overexpression and ATG7 silence respectively inhibited cell proliferation and promoted apoptosis in SH-SY5Y cells. Furthermore, we demonstrated that miR-20a-5p negatively regulated autophagy through suppressing ATG7 by binding to its 3′-UTR. These results suggested that miR-20a-5p and ATG7-mediated autophagy might play important roles in the development and progression of NB and represent as potential therapeutic targets for NB therapy.
Methods
Clinical specimens and cell lines
A total of 35 fresh NB tumor specimens were collected from Beijing Children’s Hospital, Capital Medical University between May 2015 to December 2016. Informed consent was permitted from patients/guardians upon samples collection. This study was approved by the Ethics Committees of Beijing Children’s Hospital. Histologic sections were reviewed by two expert pathologists to verify the histologic diagnosis. International Neuroblastoma Staging System (INSS) is used for clinical staging of NB by clinicians. All tissues were immediately dissected, placed on ice, and stored at liquid nitrogen until use. Neuroblastoma cell line (SH-SY5Y) and human embryonic kidney cell line (293T) were purchased from Cell Resource Center, Chinese Academy of Medical Sciences (CAMS, Beijing, China). The cells were cultured in DMEM supplemented with 10% fetal bovine serum, in a humidified, 5% CO2 incubator at 37 °C. The SH-SY5Y was chosen as a representative cell line of NB disease and is widely used in mechanism and drug development studies in NB [26, 27]. Moreover, 293T cells were employed due to their satisfactory transfection efficiency in the luciferase reporter assay.
RNA extraction and qRT-PCR assays
Tumor tissues were homogenized in TRIzol reagent (Invitrogen) and total RNA was extracted using Direct-zol™ RNA MiniPrep kit (Zymo Research) following manufacturer’s protocol. One microgram total RNA of each sample was reverse transcribed to cDNA in a final volume of 20 μl using RevertAid™ H Minus First Strand cDNA Synthesis Kit (Thermo Scientific). For microRNA reverse transcription, NCode™ miRNA First-Strand cDNA Synthesis kit (Invitrogen, USA) was used. The cDNA template was amplified using SYBR Green Master mix (Applied Biosystems, Thermo Fisher Scientific) and CFX96 Real-Time PCR System (Bio-rad). For human miRNA and mRNA expression detection, RNU6 and β-actin were used as reference genes. The primer sequences used were as follows: ATG7 (forward, 5′-TGCTATCCTGCCCTCTGTCTT-3′; reverse, 5′-TGCCTCCTTTCTGGTTCTTTT-3′); β-actin (forward, 5′-TGAGACCTTCAACACCCCAG-3′; reverse, 5′-GCCATCTCTTGCTCGAAGTC-3′); miR-20a-5 [forward, 5′-TAAAGTGCTTATAGTGCAGGTAG-3′; reverse, universal qPCR Primer (Invitrogen, USA)]; RNU6 (forward, 5′-CTCGCTTCGGCAGCACA-3′; reverse, 5′-AACGCTTCACGAATTTGCGT-3′).
Cell transfection
Oligonucleotides including miR-20a-5p mimics and miR-20a-5p inhibitor were used (Thermo Scientific, USA) for overexpression or inhibition of miR-20a-5p, respectively. For cell transfection assays, the synthetic oligonucleotides were transfected into cells using a Lipofectamine RNAiMAX Kit (Invitrogen) at about 50% confluence according to the product manual. The media were changed 8 h after transfection, and the indicated cells were subjected to further investigations.
Real-time cell proliferation assays
Real-Time cell kinetic analyzer xCELLigence RTCA (ACEA Biosciences, USA) was used to monitor dynamic changes of cell proliferation. The SH-SY5Y cells were seeded in E-plate (ACEA Biosciences, USA), and incubated at 37 °C. When the cells reached the logarithmic growth phase, the plate was detached and cells were transfected. Changes in baseline impedance resulting from the increase of cell numbers were monitored by gold micro-electrodes located at the bottom of E-plate. The proportional changes in impedance were recorded continuously and expressed as cell index (CI). The E-Plate was then incubated in the RTCA Station inside the incubator and the CI values were recorded every 20 min. Data analysis was performed using RTCA Control Unit and the preinstalled RTCA software.
Colony formation assay
Total of 1 × 103 SH-SY5Y cells were seeded in 6-well plates and transfected with oligonucleotides of mimcs or siRNA. After 10 days, cells in each well were fixed with 4% paraformaldehyde for 10 min and stained with 0.1% crystal violet for another 10 min. Cell colonies were then photographed and counted. Assays were independently conducted three times.
Caspase-Glo 3/7 assay
For the determination of caspase-3/7 activity, the cells were seeded onto 96-well plates in triplicate, and transfected as described above. At 24 h and 48 h post-transfection, caspase-3/7 activity was determined using a Caspase-Glo 3/7 kit (Promega, USA) according to the manufacturer’s protocol. Briefly, Caspase-Glo reagent was added to each well and incubated for 1 h at room temperature. Luminescence was measured using a SpectraMax Microplate Luminometer (Molecular Devices).
Luciferase reporter assays
To validate the predicted microRNA-binding sequence for miR-20a-5p, a fragment of the ATG7 3′-UTR containing either the predicted binding site or a mutated 3′-UTR was cloned into the GV272 vector (Genechem, Shanghai, China). After verification by DNA sequencing, the plasmids were transfected into 293T cells with or without synthetic miR-20a-5p mimic, using a Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Cells were collected 48 h after transfection and analyzed with the Dual-Luciferase Reporter Assay System (Promega, CA, USA). The firefly and Renilla luciferase signals were generated by SpectraMax Microplate Luminometer (Molecular Devices). Data were normalized to Renilla activity.
RNA interference
The SH-SY5Y cells in exponential growth phase were plated at 2 × 105 cells/well in 6-well plate for 24 h. Cells were then transfected with 100 nM ATG7 or scrambled siRNA using Lipofectamine RNAiMAX (Invitrogen). The siRNA oligonucleotides of ATG7 (No. 1: 5′-UCUUCGAAGUGAAGCUUCCAGAAAU-3′; No. 2: 5′-GGAGUCACAGCUCUUCCUU-3′; No. 3: 5′-CACUUCCAGUGCCUUUCCC-3′) and non-targeting control (5′-UUCUCCGAACGUGUCACGU-3′) were synthesized by Sangon Biotech, Shanghai. After transfection in antibiotic-free medium for 8 h, cells were refreshed with normal medium. Experiments were performed 72 h after transfection.
Western blotting
After SH-SY5Y cells were transfected with mimics or siRNA, the cells were lysed in RIPA lysis buffer for 30 min. Total cellular protein was extracted and determined by the bicinchoninic acid (BCA) protein assay (Pierce, USA). Equal amounts of proteins (20 μg) were separated by SDS-PAGE, and electrophoretically transferred to PVDF membranes (Millipore, USA). After blocking in 5% nonfat milk, the membranes were incubated with the following primary antibodies overnight at 4 °C: ATG7 (CST, 1:1000), LC3 (CST, 1:1000), p62 (CST, 1:1000), GAPDH (CST, 1:1000). The secondary antibody was anti-rabbit IgG (CST, 1:2000). Subsequent visualization was performed with the ECL chemiluminescence reagent (Pierce, USA), while relative densitometric analysis was performed by Image Lab software (Bio-Rad, USA).
Statistical analysis
The differences between groups were analyzed by t test or one-way analysis of variance. All data were analyzed by using the SPSS 16.0 and JMP 12.0 software. The relationship between miR-20a-5p and ATG7 mRNA was analyzed by correlation and linear regression analysis. p < 0.05 was considered to be statistically significant.
Results
Clinic-pathological characteristics of enrolled patients
A total of 35 diagnosed NB patients were recruited into our study. The summary of the clinic-pathological characteristics is detailed in Table 1, including age at diagnosis, gender, and tumor stage. More than 70% patients were less than 5 years old. According to the INSS, four patients were classified as stage I, three as stage II, nine as stage III, and nineteen as stage IV.
Both miR-20a-5p and ATG7 was dysregulated in NB samples and correlated with tumor stage
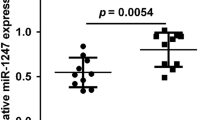
To investigate the role of miR-20a-5p and ATG7 in NB progression, quantified expression was detected in 35 NB specimens by RT-PCR. We found that the expression levels of miR-20a-5p in tumors of stage I was significantly higher than those in stage III and stage IV (Fig. 1a). However, ATG7 expression was markedly upregulated (Fig. 1b) in high staging samples compared with those in low staging samples. These results indicated that miR-20a-5p and ATG7 were significantly associated with NB stage. Moreover, correlation analysis showed that miR-20a-5p level had a negative correlation trend with ATG7 (Fig. 1c) (r = − 0.04; p = 0.632), suggesting that miR-20a-5p might involve in the regulation of ATG7.
Relative expression of miR-20a-5p and ATG7 in clinical NB tissues. a miR-20a-5p is downregulated while b ATG7 is upregulated in high staging samples compared with those in low staging samples. c Correlation analysis showed that miR-20a-5p had a negative correlation trend with ATG7 (r = − 0.04; p = 0.632)
ATG7 was a direct target of miR-20a-5p in SH-SY5Y cells
To identify the mRNA targets of miR-20a-5p, we performed a bioinformatics analysis using the publicly available algorithm (TargetScan 6.2) and found that miR-20a-5p could target and regulate ATG7 translation. To confirm the prediction, we conducted luciferase reporter assay after transfection of ATG7 3′-UTR with wild type (Wt) or mutant (Mut) into 293T cells (Fig. 2a), along with miR-20a-5p mimics or controls. As shown in Fig. 2b, miR-20a-5p reduced luciferase activity in cells transfected with Wt ATG7 3′-UTR, but no effects in cells transfected with Mut ATG7 3′-UTR (Fig. 2b). Additionally, western blotting analysis showed that miR-20a-5p dramatically suppressed ATG7 protein levels in dose- and time-dependent manners (Fig. 2c, d). These collective results suggest that miR-20a-5p negatively regulates ATG7 gene expression by directly binding to 3′-UTR of ATG7.
ATG7 was a direct target of miR-20a-5p in SH-SY5Y cells. a Schematic putative target sites of miR-20a-5p in 3′-UTR of ATG7, and the sequence of miR-20a-5p mutant (performed as mut). b Luciferase assay of wild type (WT) or mutated (Mut) ATG7 3′-UTR reporter co-transfected with miR-20a-5p or the control for 48 h. c Western blotting analysis of ATG7 protein levels in SH-SY5Y cells treated with miR-20a-5p mimics at indicated time and d concentration. The relative densitometric analysis of the protein bands were performed. *p < 0.05 vs. control. Data are expressed as mean ± SD, n = 3. miR-NC, negative control microRNA
MiR-20a-5p overexpression and ATG7 knockdown suppressed proliferation and progression in SH-SY5Y cells
To investigate biological function of miR-20a-5p in proliferation of neuroblastoma, miR-20a-5p mimics were transfected into SH-SY5Y cells and RTCA device was used to monitor cell index in real time (Fig. 3a). Quantitative analysis result showed that miR-20a-5p overexpression significantly inhibited the proliferation activity of SH-SY5Y cells (Fig. 3b). Moreover, colony formation assay further revealed that miR-20a-5p markedly suppressed SH-SY5Y cell growth, as indicated by reduction of colony numbers and intensity (Fig. 3c). These results indicated that miR-20a-5p overexpression suppressed proliferation and progression of SH-SY5Y cells.
MiR-20a-5p inhibited cell proliferation and promotes cellular apoptosis in SH-SY5Y cells. a Effects of miR-20a-5p overexpression on cell proliferation measured by Real-Time cell kinetic analyzer (RTCA) and b quantification. c Representative photographs of colonies formed in 14 days after mimics transfection. d Caspase-3/7 activity in SH-SY5Y cells treated with miR-20a-5p mimics at indicated time and concentration. Data are expressed as mean ± SD, n = 3. *p < 0.05 vs. control, Student’s t test. miR-NC, negative control microRNA
To evaluate the effects of ATG7 on SH-SY5Y cells progression, we suppressed endogenous ATG7 expression by specific siRNAs. As shown in Fig. 4a, ATG7-targeted siRNA effectively silenced ATG7 expression (Fig. 4a), and significantly inhibited proliferation of SH-SY5Y cells both in RTCA real-time detection and quantitative analysis (Fig. 4b, c). The colony formation assay indicated that ATG7 knockdown suppressed colony forming ability in SH-SY5Y cells (Fig. 4d). These results indicated that ATG7 knockdown suppressed proliferation and progression in SH-SY5Y cells.
ATG7 inhibited cell proliferation and promotes cellular apoptosis in SH-SY5Y cells. a Western blotting validation of siRNAs on ATG7 knockdown. b Effects of ATG7 knockdown on cell proliferation measured by Real-Time cell kinetic analyzer (RTCA) and c quantification. d Representative photographs of colonies formed in 10 days after siRNA1 transfection. e Caspase-3/7 activity in SH-SY5Y cells transfected with ATG7 siRNA1 and siRNA2. Data are expressed as mean ± SD, n = 3. *p < 0.05 vs. control, Student’s t test. siRNA-NC, negative control siRNA
MiR-20a-5p overexpression and ATG7 knockdown suppressed autophagy in SH-SY5Y cells
To investigate the suppression of miR-20a-5p on autophagy, rapamycin was applied to establish autophagy model in SH-SY5Y cells. In circumstances of autophagy induced by rapamycin (Fig. 5a), relative densitometric analysis showed that miR-20a-5p inhibited autophagy marker of LC3-II/LC3-I ratio and ATG7 expression. In addition, miR-20a-5p inhibitor significantly reversed these effects. Thus miR-20a-5p was demonstrated to suppress autophagy and ATG7 expression (Fig. 5b). Since ATG7 was identified as a direct target of miR-20a-5p in luciferase reporter assay, these results clarified that ATG7 potentially acted as an executor underlying miR-20a-5p-mediated autophagy suppression.
Effects of miR-20a-5p and ATG7 on autophagy in SH-SY5Y cells. a Western blotting validation of ATG7 knockdown on autophagy suppression and b relative densitometric analysis of the protein bands. c Forced miR-20a-5p expression inhibited ATG7 expression and suppressed rapamycin-induced autophagy, while miR-20a-5p inhibitor reversed these effects. d Relative densitometric analysis of the protein bands. *p < 0.05 vs. indicated groups. Data are expressed as mean ± SD, n = 3. Rapa, rapamycin; miR-NC, negative control microRNA
Since ATG7 plays an essential role in complete autophagy, the effects of ATG7 knockdown on autophagy inhibition was further verified by measuring LC3-II/LC3-I ratio (autophagy marker) and autophagy substrate SQSTM1/p62. As shown in western blotting (Fig. 5c), all the ATG7 specific siRNAs increased LC3-I levels, suggesting that the conversion of LC3-I to LC3-II was inhibited. The elevated LC3-I thus accounts for significantly decrease of LC3-II/LC3-I ratio (Fig. 5d). Since p62 is a substrate for autophagy degradation, increased p62 expression (Fig. 5c) by ATG7-targeted siRNA confirmed that ATG7 knockdown suppressed cellular autophagy. These results validated that ATG7 mediated autophagy activation in SH-SY5Y cells.
MiR-20a-5p overexpression and ATG7 knockdown increased caspase-3/7 activity
To determine the effects of miR-20a-5p and ATG7 on cellular apoptosis, caspase-3/7 activity in SH-SY5Y cells were examined post-transfection with miR-20a-5p mimics and ATG7 siRNA, respectively. The results showed that caspase-3/7 activity was significantly increased in SH-SY5Y cells transfected with miR-20a-5p mimics in a time- and dose-dependent manner, as compared with the negative control (Fig. 3d). Similarly, ATG7 siRNA significantly enhanced the activity of caspase-3/7 in SH-SY5Y cells (Fig. 4e). These results suggested that miR-20a-5p overexpression and ATG7 knockdown promoted cellular apoptosis, respectively.
Discussion
Neuroblastoma (NB) is the most common extracranial solid tumor in children. Despite receiving multimodal treatment, high-risk NB remains a clinical challenge with survival rates below 50% [2]. Even though genetic alterations have been identified, such as MYCN, ALK, ATRX and TERT [3,4,5], mechanisms underlying NB progression were still not clear. In the present study, we found that miR-20a-5p was downregulated while ATG7 was upregulated in NB tumor specimens. Additional bioinformatics analysis suggested that miR-20a-5p could bind to 3′-UTR of ATG7 and suppress ATG7 translation. Further evidence confirmed that miR-20a-5p inhibited cell proliferation and promoted apoptosis through ATG7-mediated autophagy in SH-SY5Y cells (Fig. 6).
The putative schematic representation of pathway involved in miR-20a-5p-mediated suppression of cell proliferation in SH-SY5Y cells. MiR-20a-5p recognizes and binds to 3′-UTR of ATG7, repressing ATG7 gene translation. Since ATG7 contribute to autophagosome initiation and completion, which accounts for cell proliferation and anti-apoptosis, miR-20a-5p-mediated ATG7 inhibition thus results in cell proliferation suppression and further apoptosis
MiRNAs are involved in the etiology, progression, and prognosis of cancer [7, 9]. For tumorigenesis, miRNAs produce competing oncogenic and tumor suppressive effects by suppressing both tumor suppressive mRNAs and oncogenic mRNAs, respectively [9, 28, 29]. To verify the effects of miR-20a-5p on NB tumorigenesis, in vitro experiments were performed in this study, demonstrating that miR-20a-5p inhibited cell proliferation and colony formation. Consistent with our findings that miR-20a-5p was downregulated in pediatric NB, miR-451 expression was reported reduced in NB specimens, and correlated with tumour size, lymph node and distant metastasis, and tumour-node-metastasis (TNM) stage [13].
Functionally, miRNAs regulate gene expression by suppressing target mRNA translation and reducing mRNA stability. Herein, autophagy-related gene 7 (ATG7) was a bioinformatics target of miR-20a-5p, and luciferase assay validated miR-binding sequence for miR-20a-5p. Western blotting analysis further showed that forced expression of miR-20a-5p dramatically suppressed ATG7 protein levels in a dose- and time-dependent manner. Since ATG7 is crucial in autophagy activation and autophagosome formation [30], these results suggest that miR-20a-5p may inhibit cell proliferation through autophagy via negative regulation of ATG7 in SH-SY5Y cells. Similarly, miR-337-3p was reported to suppress NB progression by repressing the transcription of matrix metalloproteinase 14 [31]. However, further study was needed to clarify that ATG7-mediated autophagy contributed to NB progression.
The degradation process of autophagy is controlled by autophagy-related genes (ATGs), including ATG7 [32]. Normally, autophagy plays an essential role in maintaining cellular homeostasis and physiological functions. However, autophagy confers stress tolerance as a carcinogenetic effect, which promote tumor development, malignant transformation and drug or apoptosis resistance [21, 33, 34]. In clinical NB tissues, we found that ATG7 was upregulated. In an in vitro model, ATG7 knockdown significantly inhibited cellular proliferation and colony formation as well, suggesting that ATG7-mediated autophagy may be involved in cell proliferation and tumor growth of pediatric NB. Consistent with our findings, ATG7 knockout can significantly reduce tumor cell tumorigenicity and promote the transformation of lung cancer into benign tumor [35]. In addition, ATG7 deficiency can completely inhibit tumor occurrence and development of intestinal epithelial cells in mice [36]. Herein, ATG7 knockdown was further validated to inhibit autophagy induction and substrate degradation of SQSTM1/p62. Taken all these results into consideration, ATG7-mediated autophagy is was postulated to contribute to NB progression.
In the present study, miR-20a-5p was demonstrated to suppress ATG7 expression and autophagy, while miR-20a-5p inhibitor reversed these effects. Such evidences indicated the involvement of miR-20a-5p in autophagy. Previously, miRNAs were reported to account for the regulation of autophagy, including autophagy initiation, autophagy vesicles formation and substrates degradation [17]. Numerous miRNAs regulate autophagy by suppressing autophagy-related proteins, such as miR-17, miR-20a, miR-106b and miR-375 [22, 37, 38]. Wu et al. reported that miR-20a and miR-106b negatively regulate starvation-induced autophagy by suppressing ULK1 in C2C12 myoblasts [39]. Due to the fact that ATG7 can simultaneously regulate two ubiquitin-like systems of ATG12 and ATG8 in autophagy activation and autophagosome formation [30], the miR-20a-5p-mediated silence of ATG7 indicated that miR-20a-5p contributed to autophagy inhibition.
There also exists crosstalk between autophagy and apoptosis even though the details of molecular switching points are still not fully elucidated [40, 41]. Autophagy inhibition by ATG7 silence significantly enhanced the activity of caspase-3/7, suggesting that suppression of autophagy promotes cellular apoptosis. In line with our results, knockdown of essential autophagy genes in tumor cells could also potentiate the induction of cell death. In human pancreatic cancer, inhibition of autophagy led to tumor regression and extended survival in xenografts and genetic mouse models [42]. In the present study, miR-20a-5p suppressed autophagy and ATG7 expression and enhanced cellular apoptosis, indicating that autophagy might contribute to cell proliferation and apoptosis resistance in SH-SY5Y cells. The putative molecular mechanism underlying miR-20a-5p-mediated effects in SH-SY5Y cells is presented in the schematic diagram. This study improved our understanding that miRNAs-mediated autophagy was involved in the regulation of NB development and may provide potential new therapeutic targets for the management of pediatric NB.
Conclusions
In summary, we demonstrated that miR-20a-5p was significantly downregulated while ATG7 was upregulated along with clinical staging of NB progression. Both miR-20a-5p overexpression and ATG7 silence impaired cell growth, colony formation and promoted apoptosis in SH-SY5Y cells in vitro. Furthermore, we confirmed that miR-20a-5p negatively regulated autophagy through suppressing ATG7 by binding to its 3′-UTR. This study extends our knowledge about ATG7 regulation by miRNAs, and suggests that both miR-20a-5p and ATG7 may be of potential value as novel therapeutic targets for pediatric NB.
Abbreviations
- NB:
-
neuroblastoma
- ATG7:
-
autophagy-related gene 7
- UTR:
-
untranslated region
- miRNA:
-
microRNA
- FBS:
-
fetal bovine serum
- RNU6B:
-
U6B small nuclear RNA
References
Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, et al. Neuroblastoma. Nat Rev Dis Primers. 2016;2:16078.
Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–20.
Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–84.
Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–93.
Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526:700–4.
Bartholomew J, Washington T, Bergeron S, Nielson D, Saggio J, Quirk L. Dinutuximab: a novel immunotherapy in the treatment of pediatric patients with high-risk neuroblastoma. J Pediatr Oncol Nurs. 2016.https://doi.org/10.1177/1043454216659448.
Berindan-Neagoe I, Monroig PD, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. Ca-Cancer J Clin. 2014;64:311–36.
Iwakawa HO, Tomari Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015;25:651–65.
Lin SB, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–33.
Cheng L, Yang T, Kuang Y, Kong B, Yu S, Shu H, et al. MicroRNA-23a promotes neuroblastoma cell metastasis by targeting CDH1. Oncol Lett. 2014;7:839–45.
Li Y, Li W, Zhang JG, Li HY, Li YM. Downregulation of tumor suppressor menin by miR-421 promotes proliferation and migration of neuroblastoma. Tumour Biol. 2014;35:10011–7.
Qu HX, Zheng LD, Pu JR, Mei H, Xiang X, Zhao X, et al. miRNA-558 promotes tumorigenesis and aggressiveness of neuroblastoma cells through activating the transcription of heparanase. Hum Mol Genet. 2015;24:2539–51.
Liu G, Xu Z, Hao D. MicroRNA451 inhibits neuroblastoma proliferation, invasion and migration by targeting macrophage migration inhibitory factor. Mol Med Rep. 2016;13:2253–60.
Huang D, Bian G, Pan Y, Han X, Sun Y, Wang Y, et al. MiR-20a-5p promotes radio-resistance by targeting Rab27B in nasopharyngeal cancer cells. Cancer Cell Int. 2017;17:32.
Cheng D, Zhao S, Tang H, Zhang D, Sun H, Yu F, et al. MicroRNA-20a-5p promotes colorectal cancer invasion and metastasis by downregulating Smad4. Oncotarget. 2016;7:45199–213.
Pu Y, Yi Q, Zhao F, Wang H, Cai W, Cai S. MiR-20a-5p represses multi-drug resistance in osteosarcoma by targeting the KIF26B gene. Cancer Cell Int. 2016;16:64.
Su Z, Yang Z, Xu Y, Chen Y, Yu Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget. 2015;6:8474–90.
Lozy F, Karantza V. Autophagy and cancer cell metabolism. Semin Cell Dev Biol. 2012;23:395–401.
Nixon RA, Yang D-S. Autophagy and neuronal cell death in neurological disorders. Cold Spring Harb Perspect Biol. 2012;4:a008839.
Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–62.
Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34:856–80.
Chang Y, Yan W, He X, Zhang L, Li C, Huang H, et al. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. 2012;143(177–87):e8.
Pan B, Feng B, Chen Y, Huang G, Wang R, Chen L, et al. MiR-200b regulates autophagy associated with chemoresistance in human lung adenocarcinoma. Oncotarget. 2015;6:32805–20.
Belounis A, Nyalendo C, Le Gall R, Imbriglio TV, Mahma M, Teira P, et al. Autophagy is associated with chemoresistance in neuroblastoma. BMC Cancer. 2016;16:891.
Liu Y, Song L. HMGB1-induced autophagy in Schwann cells promotes neuroblastoma proliferation. Int J Clin Exp Pathol. 2015;8:504–10.
Li Z, Yan S, Attayan N, Ramalingam S, Thiele CJ. Combination of an allosteric Akt inhibitor MK-2206 with etoposide or rapamycin enhances the antitumor growth effect in neuroblastoma. Clin Cancer Res. 2012;18:3603–15.
Radogna F, Cerella C, Gaigneaux A, Christov C, Dicato M, Diederich M. Cell type-dependent ROS and mitophagy response leads to apoptosis or necroptosis in neuroblastoma. Oncogene. 2016;35:3839–53.
Oberg K, Modlin IM, De Herder W, Pavel M, Klimstra D, Frilling A, et al. Consensus on biomarkers for neuroendocrine tumour disease. Lancet Oncol. 2015;16:e435–46.
Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor suppressor? The duplicity of microRNAs in cancer. Cancer Res. 2016;76:3666–70.
Das G, Shravage BV, Baehrecke EH. Regulation and function of autophagy during cell survival and cell death. Cold Spring Harb Perspect Biol. 2012;4:a008813.
Xiang X, Mei H, Zhao X, Pu J, Li D, Qu H, et al. miRNA-337-3p suppresses neuroblastoma progression by repressing the transcription of matrix metalloproteinase 14. Oncotarget. 2015;6:22452–66.
Galluzzi L, Bravo-San Pedro JM, Vitale I, Aaronson SA, Abrams JM, Adam D, et al. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differ. 2015;22:58–73.
Jiang X, Overholtzer M, Thompson CB. Autophagy in cellular metabolism and cancer. J Clin Investig. 2015;125:47–54.
White E, Mehnert JM, Chan CS. Autophagy, metabolism, and cancer. Clin Cancer Res. 2015;21:5037–46.
Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3:1272–85.
Levy J, Cacheux W, Bara MA, L’Hermitte A, Lepage P, Fraudeau M, et al. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat Cell Biol. 2015;17:1062–73.
Comincini S, Allavena G, Palumbo S, Morini M, Durando F, Angeletti F, et al. microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol Ther. 2013;14:574–86.
Wang IK, Sun KT, Tsai TH, Chen CW, Chang SS, Yu TM, et al. MiR-20a-5p mediates hypoxia-induced autophagy by targeting ATG16L1 in ischemic kidney injury. Life Sci. 2015;136:133–41.
Wu H, Wang F, Hu S, Yin C, Li X, Zhao S, et al. MiR-20a and miR-106b negatively regulate autophagy induced by leucine deprivation via suppression of ULK1 expression in C2C12 myoblasts. Cell Signal. 2012;24:2179–86.
Green DR, Galluzzi L, Kroemer G. Metabolic control of cell death. Science. 2014;345:1466.
Lalaoui N, Lindqvist LM, Sandow JJ, Ekert PG. The molecular relationships between apoptosis, autophagy and necroptosis. Semin Cell Dev Biol. 2015;39:63–9.
Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29.
Authors’ contributions
XN and YLG designed the experiment and critically revised the paper; YBY performed most of the experiments and wrote the manuscript; JZ and HMW contributed to the patient recruitment and tumor tissue collection; YQJ, YRY and JL performed the bioinformatic prediction; JS and FC cultured cells; SJH and PC conducted statistical analysis. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare they have no competing interests.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Consent for publication
Informed consent was obtained from all individual participants included in the study.
Ethics approval and consent to participate
This study was approved by the Ethics Committees of Beijing Children’s Hospital and prior informed consent was obtained from all patients.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 81702463, 81472369, 81502144), Beijing Health System Top Level Technical Personnel Training Plan (No. 20153079) and Beijing Children’s Hospital Young Investigator Program (BCHYIPB-2016-08).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yu, Y., Zhang, J., Jin, Y. et al. MiR-20a-5p suppresses tumor proliferation by targeting autophagy-related gene 7 in neuroblastoma. Cancer Cell Int 18, 5 (2018). https://doi.org/10.1186/s12935-017-0499-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-017-0499-2