Abstract
Background
Escherichia coli is an important strain for l-threonine production. Genetic switch is a ubiquitous regulatory tool for gene expression in prokaryotic cells. To sense and regulate intracellular or extracellular chemicals, bacteria evolve a variety of transcription factors. The key enzymes required for l-threonine biosynthesis in E. coli are encoded by the thr operon. The thr operon could coordinate expression of these genes when l-threonine is in short supply in the cell.
Results
The thrL leader regulatory elements were applied to regulate the expression of genes iclR, arcA, cpxR, gadE, fadR and pykF, while the threonine-activating promoters PcysH, PcysJ and PcysD were applied to regulate the expression of gene aspC, resulting in the increase of l-threonine production in an l-threonine producing E. coli strain TWF001. Firstly, different parts of the regulator thrL were inserted in the iclR regulator region in TWF001, and the best resulting strain TWF063 produced 16.34 g l-threonine from 40 g glucose after 30 h cultivation. Secondly, the gene aspC following different threonine-activating promoters was inserted into the chromosome of TWF063, and the best resulting strain TWF066 produced 17.56 g l-threonine from 40 g glucose after 30 h cultivation. Thirdly, the effect of expression regulation of arcA, cpxR, gadE, pykF and fadR was individually investigated on l-threonine production in TWF001. Finally, using TWF066 as the starting strain, the expression of genes arcA, cpxR, gadE, pykF and fadR was regulated individually or in combination to obtain the best strain for l-threonine production. The resulting strain TWF083, in which the expression of seven genes (iclR, aspC, arcA, cpxR, gadE, pykF, fadR and aspC) was regulated, produced 18.76 g l-threonine from 30 g glucose, 26.50 g l-threonine from 40 g glucose, or 26.93 g l-threonine from 50 g glucose after 30 h cultivation. In 48 h fed-batch fermentation, TWF083 could produce 116.62 g/L l‐threonine with a yield of 0.486 g/g glucose and productivity of 2.43 g/L/h.
Conclusion
The genetic engineering through the expression regulation of key genes is a better strategy than simple deletion of these genes to improve l-threonine production in E. coli. This strategy has little effect on the intracellular metabolism in the early stage of the growth but could increase l-threonine biosynthesis in the late stage.
Similar content being viewed by others
Introduction
l-Threonine has an increasing market demand in pharmaceutical, cosmetics and food industry [1]. Escherichia coli is an important strain for l-threonine production [2,3,4]. Efforts have been made to improve the productivity of l-threonine in E. coli, such as increasing the carbon flux to l-threonine [5, 6], enhancing the export of l-threonine [7], weakening the competitive pathway and reducing the consumption of l-threonine [8, 9].
Genetic switch is a ubiquitous regulatory tool for gene expression in prokaryotic cells [10]. In E. coli, l-threonine is synthesized through a series of reactions from l-aspartate, which is converted from oxaloacetate by aspartate aminotransferase encoded by aspC. The key enzymes required for l-threonine biosynthesis in E. coli are encoded by the thr operon. The thr operon could coordinate expression of these genes when l-threonine is in short supply in the cell. This regulation is controlled by the lead sequence of the thr operon (thrL). The thrL contains an efficient ribosome-binding site (RBS) and can form a twenty-one-amino-acid leader peptide. From the 6th codons of this leader peptide, it starts to encode l-threonine residues, therefore, it can regulate the transcription of the thr operon through the availability of l-threonine, by forming different base-paired RNA structure [11]. Since thrL inhibits l-threonine accumulation it has been mutated in l-threonine producing E. coli, such as TWF001 [9]. On the other hand, some biosynthetic pathways in E. coli can be activated in the presence of high concentration of l-threonine, such as the sulfate metabolism branch of the cysteine biosynthetic pathway which includes the genes cysDN, cysJI and cysH [12].
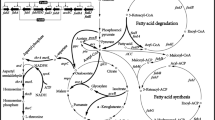
To sense and regulate intracellular or extracellular chemicals, bacteria evolve a variety of transcription factors. Based on this principle, various biosensors for amino acids and their precursors, including l-methionine, l-leucine, l-isoleucine, l-valine [13, 14], l-lysine, l-arginine, l-serine, O-acetyl-l-serine [15], O-acetyl homoserine [16], and oxygen [17] have been constructed, and some of these sensors have been successfully applied to the high-throughput screening [12, 18, 19] and systems metabolic engineering [20]. l-Threonine production in E. coli can be enhanced by regulating IclR [8, 9], a transcription factor. IclR is a repressor for the aceBAK operon (Fig. 1), which encodes the three enzymes in the glyoxylate bypass [21]. The deletion of iclR in E. coli can improve production of phloroglucinol and 3-hydroxypropionate [22], produce succinate from acetate [23], and produce fumaric acid [24, 25], ectoine [26] and fatty acids [27]. The metabolism regulator protein FadR represses the transcription of the fadBA operon, fadE, fadD, fadH, fadIJ operon, fadL and fadM, which involved in the fatty acid degradation [28]. FadR can also activate the express of gene iclR and the genes accA, accD, accBC operon, fabA, fabB and fabHDG operon involved in the fatty acid biosynthesis (Fig. 1). Therefore, the absence of FadR can weaken the lipid biosynthesis and enhance the fatty acid degradation in E. coli [29,30,31]. The iclR deletion mutant strain TWF006 further modified by enhancing fatty acid degradation and the glyoxylate shunt can increase l-threonine production [6]. CpxR and GadE can also activate the genes involved in the dissociated or type II fatty acid synthase systems, such as fabA and fabZ [32, 33]. Under the anaerobic conditions, ArcA represses the genes involved in TCA cycle and glyoxylate shunt [34], such as gltA [35], acnAB [36], icd [37], sucABCD [38], lpd [39], sdhCDAB [40], fumA, fumC [41], fumB [42], mdh [43], maeA and aceAB [44], and the key genes aceEF and lpdA [45] which encoding the enzymes reaction from pyruvate to acetyl-CoA. The l-threonine production in E. coli strain TWF001 was increased when arcA and iclR were deleted [46]. Expressing two pyruvate kinase isoenzymes, PYKI and PYKII, encoded by pykF and pykA, respectively, is important for cellular metabolism and glycolysis in E. coli. In pykF deletion mutant, the flux of Embden-Meyerhof-Parnas pathway was reduced from 65 to 20% while the flux of pentose phosphate pathway was increased from 34 to 79% [47]. Acetate, the main by-product of E. coli at high cell density, can inhibit the cell growth and l-threonine production. Since acetate formation is correlated with the metabolic flux overflow, pykF deletion can lead to less acetate and more l-threonine production in an l-threonine-producing E. coli THRD [48]. In this study, l-threonine was used to regulate the expression of genes iclR, arcA, cpxR, gadE, fadR, pykF, and aspC through thrL and the promoters of cysD, cysH and cysJ, leading to the increased l-threonine production in E. coli TWF001 (Fig. 1). The expression of these seven genes in E. coli are controlled by the concentration of l-threonine. The final strain TWF083 constructed in this study could produce 26.50 g/L l-threonine after 21 h flask fermentation, which was 49.4% increase compared to the control strain TWF001.
The glycolysis, TCA cycle, fatty acid metabolism, l-threonine biosynthetic pathway, and their regulation in E. coli. Genes encoding for the corresponding metabolic enzymes are shown in italic. The genes regulated by IclR, ArcA, FadR, GadE and CpxR are labeled by the different symbols. GLU glucose, G6P glucose-6-phosphate, PEP phosphoenolpyruvate, PYR pyruvate, AcCoA acetyl-coA, CIT citrate, ICT isocitrate, α-KG α-ketoglutarate, SUC-CoA succinyl-CoA, SUC succinate, FUM fumarate, MAL malate, OAA oxaloacetate, ASP aspartate, THR threonine, FOR formate, Acy-ACP Acyl-ACP, En-ACP Enoyl-ACP, 3Ha-ACP 3-Hydroxyacyl-ACP, 3Ka-ACP 3-Ketoacyl-ACP, MaCoA Malonyl-CoA, 3HaCoA 3-Hydroxyacyl-CoA, EnCoA Enoyl-CoA, AcyCoA Acyl-CoA, 3KaCoA 3-Ketoacyl-CoA
Materials and method
Chemicals and reagents
Plasmids and genomic DNA were extracted using the Plasmids mini kit and DNA purification kit (Tiangen, Beijing, China), respectively. PrimerSTAR HS DNA polymerase (Takara, Dalian, China) was used for PCR. Restriction endonucleases and T4 DNA ligase were purchased from Thermo Scientific (Waltham, USA). Total RNA was extracted using RNA extraction kit (Bio Flux, Beijing, China). The RNA was reversely transcribed into cDNA using the Revert Aid™ First Strand cDNA synthesis kit (Fermentas, Shanghai, China). RT-PCR was performed using the Real Master Mix kit (Tiangen, Beijing, China). The intracellular concentration of NADPH/NADP+ was determined by using NADPH/NADP+ Kit (Beyotime Biotechnology, Shanghai, China).
Bacterial growth condition
The bacterial strains and plasmids used in this study are listed in Table 1. The primers used in this study are listed in Table 2. The promoters used in this study are listed in Table 3. The ribosome binding sites (RBSs) used in this study are listed in Table 4. The Luria–Bertani (LB) medium (5 g/L yeast extract, 10 g/L tryptone, and 10 g/L NaCl) was used to culture the E. coli cells at 37 °C or 30 °C with 200 rpm shaking. To maintain the plasmids in the engineered strains or to provide the selective pressure, kanamycin (50 mg/L), spectinomycin (50 mg/L), isopropyl-d-thiogalactopyranoside (IPTG; 0.5 mM) or arabinose (10 mM) was supplemented in the medium.
Genome sequence of E. coli MG1655 was used to design the primer sequences because the strain TWF001 [9] was derived from MG1655. The thr operon regulatory region was amplified using the genome sequence of MG1655 as template because the sequence of thr operon regulatory region in TWF001 was mutated. The insertion fragments were amplified by obtaining the insertion sequence in the first PCR, and then adding the homologous arm in the second PCR. The database on https://salislab.net was used to the design ribosome binding site (RBS) sequence and calculate its translation initiation rate.
Construction of E. coli mutant strains TWF051, TWF052, TWF053, TWF054 and TWF055
The RBS sequence of the five genes arcA, cpxR, gadE, pykF and fadR in TWF001 was individually replaced with thrR (RBSthrL-thrL-RBSthrA, the combined sequence of the RBS sequence of thrL, thrL, and the RBS sequence of thrA). As shown in Fig. 2a, TWF051 was derived from TWF001 by replacing the native RBS of arcA in chromosome with the thrR-arcA fragments (the thrR fragment with the homologous arm of arcA), using the CRISPR-Cas9 two-plasmid system [49]. Firstly, the fragments of thrR-arcA were amplified using the primer pairs ParcA-U-FR1/ParcA-D-RF1and using the genomic DNA of E. coli MG1655 as the template, then amplified using the primer pairs ParcA-U-FR11/ParcA-D-RF11and using the recovered fragments as a template. Secondly, the plasmid pTargetF-arcA was constructed though inverse PCR using the primers Pf-ParcA-F/pf-ParcA-R and using pTargetF as the template, and then self-ligation; the primers Pf-ParcA-V-F/pf-V-R was used to confirm the correctness of the plasmid by PCR. Next, pTargetF-arcA and thrR-arcA were used according to the instruction of the CRISPR–Cas9 system [49]. After 36 h culturing at 30 °C, the correctness of strain was identified using the primer pairs ThrL-V-F/ParcA-V-R, and sequenced using the primers ParcA-S-F/ParcA-S-R. Using the same method, TWF052 was derived from TWF001 by replacing the native RBS of cpxR in chromosome with the thrR-cpxR fragments. The fragment of thrR-cpxR was amplified using the primer pairs PcpxR-U-FR/PcpxR-D-RF, and then using the primer pairs PcpxR-U-FR11/PcpxR-D-RF11. The plasmid pTargetF-cpxR was constructed using the primers Pf-PcpxR-F/pf-PcpxR-R, and the primers Pf-PcpxR-V-F/pf-V-R was used to confirm the correctness of the plasmid by PCR. The correctness of strain was identified using the primer pairs ThrL-V-F/PcpxR-V-R, and sequenced using the primers PcpxR-S-F/PcpxR-S-R. TWF053 was derived from TWF001 by replacing the native RBS of cpxR in chromosome with the thrR-gadE fragments. The fragments of thrR-gadE were amplified using PgadE-U-FR/PgadE-D-RF, and then using PgadE-U-FR11/PgadE-D-RF11. The plasmid pTargetF-gadE was constructed using the primers Pf-PgadE-F/pf-PgadE-R and confirmed using the primers Pf-PgadE-V-F/pf-V-R. The primer pairs ThrL-V-F/PgadE-V-R and the primers PgadE-S-F/PgadR-S-R were used to identify and sequence the correctness of strain, respectively. TWF054 was derived from TWF001 by replacing the native RBS of pykF in chromosome with the thrR-gadE fragments. The fragments of thrR-pykF were amplified using PpykF-U-FR/PpykF-D-RF, and then using PpykF-U-FR11/PpykF-D-RF11. The plasmid pTargetF-pykF was constructed using the primers Pf-PpykF-F/pf-PpykF-R and confirmed using the primers Pf-PpykF-V-F/pf-V-R. The primer pairs ThrL-V-F/PpykF-V-R and the primers PpykF-S-F/PpykF-S-R were used to identify and sequence the correctness of strain. TWF055 was derived from TWF001 by replacing the native RBS of fadR in chromosome with the thrR-fadR fragments. The fragments of thrR-fadR were amplified using PfadR-U-FR/PfadR-D-RF, and then using PfadR-U-FR11/PfadR-D-RF11. The plasmid pTargetF-fadR was constructed using the primers Pf-PfadR-F/pf-PfadR-R and confirmed using the primers Pf-PfadR-V-F/pf-V-R. The primer pairs ThrL-V-F/PfadR-V-R and the primers PfadR-S-F/PfadR-S-R were used to identify and sequence the correctness of strain, respectively.
Flask cultivation for l-threonine production in E. coli strains TWF001, TWF0010, TWF051, TWF052, TWF053, TWF054 and TWF055. a Genetic comparison of the strains constructed by dynamic regulation genes arcA, cpxR, gadE, pykF and fadR. b Cell growth; c glucose consumption; dl-threonine production; e relative transcription levels of arcA, cpxR, gadE, pykF and fadR in TWF051, TWF052, TWF053, TWF054 and TWF055, respectively. The error bars indicate the standard deviations from three independent experiments
Construction of E. coli mutant strains TWF056, TWF057, TWF058, TWF059, TWF060, TWF061, TWF062, TWF063, TWF064 and TWF065
As shown in Fig. 3a, TWF056 was derived from TWF001 by inserting the thrL-RBSthrA (thrR1) fragment between the native ribosomal binding site of the iclR (RBSiclR) and the iclR sequence, using the CRISPR–Cas9 two-plasmid system [49]. Firstly, the thrR1 fragment was amplified using the primer pairs PiclR-U-FR1/PiclR-D-RF1 and using the genomic DNA of MG1655 as a template, and then amplified using the primer pairs PiclR-U-FR11/PiclR-D-RF1 and using the recovered fragments as a template. Secondly, the plasmid pTargetF-iclR1 was constructed using the primers Pf-PiclR-F/pf-PiclR-R by inverse PCR using pTargetF as template, and then self-ligation. The primers Pf-PiclR-V-F/pf-V-R was using to confirm the correctness of the plasmid by PCR. The primer pairs ThrL-V-F/PiclR-V-R and the primers PiclR-S-F/PiclR-S-R were used to identify and sequence the correctness of strain, respectively. Using the same method, TWF057 was constructed. RBSthrL-thrL (thrR2) was amplified using the primers PiclR-U-FR2/PiclR-D-RF2, and then amplified using the PiclR-U-FR22/PiclR-D-RF22. The plasmid pTargetF-iclR2 was constructed using the primers Pf-PiclR-F2/pf-PiclR-R2, and identified using the primers Pf-PiclR-V-F2/pf-V-R). To construct TWF058, the sequence RBSthrL-thrL-RBSthrA (thrR) was amplified using PiclR-U-FR2/PiclR-D-RF1, and then amplified using PiclR-U-FR22/PiclR-D-RF11. The plasmid pTargetF-iclR1 was used. To constructed TWF059, the sequence PthrL-thrR was amplified using PiclR-U-FR3/PiclR-D-RF1, and then amplified using PiclR-U-FR33/PiclR-D-RF11. The plasmid pTargetF-iclR1 was used. To construct TWF060, the sequence RBSthrL-thrL-RBSs1 (thrRs1) was amplified using PiclR-U-FR2/PiclR-D-RF4, and then amplified using PiclR-U-FR22/PiclR-D-RF44. The plasmid pTargetF-iclR1 was used. To construct TWF061, the sequence RBSs2-thrL-RBSthrA (thrRs2) was amplified using PiclR-U-FR5/PiclR-D-RF4, and then amplified using PiclR-U-FR55/PiclR-D-RF44. The plasmid pTargetF-iclR1 was used. To construct TWF062, the sequence RBSs3-thrL-RBSthrA (thrRs3) was amplified using PiclR-U-FR6/PiclR-D-RF4, and then amplified using PiclR-U-FR55/PiclR-D-RF44. The plasmid pTargetF-iclR1 was used. To construct TWF063, the sequence RBSs4-thrL-RBSthrA (thrRs4) was amplified using PiclR-U-FR7/PiclR-D-RF4, and then amplified using PiclR-U-FR55/PiclR-D-RF44. The plasmid pTargetF-iclR1 was used. To construct TWF064, the sequence RBSs5-thrL-RBSthrA (thrRs5) was amplified using PiclR-U-FR8/PiclR-D-RF4, and then amplified using PiclR-U-FR55/PiclR-D-RF44. The plasmid pTargetF-iclR1 was used. To construct TWF065, the sequence RBSs6-thrL-RBSthrA (thrRs6) was amplified using PiclR-U-FR9/PiclR-D-RF4, and then amplified using PiclR-U-FR55/PiclR-D-RF44. The plasmid pTargetF-iclR1 was used.
Flask cultivation for l-threonine production in E. coli strains TWF001, TWF003, TWF056, TWF057, TWF058, TWF059, TWF060, TWF061, TWF062, TWF063, TWF064, TWF065. a Genetic comparison of the strains constructed by dynamic regulation genes iclR. b Cell growth; c glucose consumption; dl-threonine production; e relative transcription levels of iclR. The error bars indicate the standard deviations from three independent experiments
Construction of E. coli mutant strains TWF066, TWF067, TWF068, TWF069, TWF070, TWF071, TWF072, TWF073, TWF074, TWF075, TWF076, TWF077, TWF078, TWF079, TWF080, TWF081, TWF082 and TWF083
As shown in Fig. 4a, TWF066 was derived from TWF063 by inserting the gene aspC controlled by the promoter of cysH (PcysH) and RBSs7 into the lacI locus in chromosome. The fragment of RBSs7-aspC was amplified by using the primers cysH-aspC-F2/cysH-aspC-R2 and using the genomic DNA of TWF001 as a template. The promoter of cysH was amplified by using the primers cysH-aspC-F1/cysH-aspC-R1 and using the genomic DNA of TWF001 as template. These two DNA fragments were linked together by overlap PCR, using the primers cys-aspC-F/cys-aspC-R, and resulting in the replacement fragment PcysH-RBSs7-aspC. The plasmid pTargetF-lacI was constructed using Pf-lacI-F/pf-lacI-R, and confirmed using Pf-lacI-V-F/pf-V-R. The primers lacI-aspC-V-F/lacI-aspC-V-R and lacI-S-F/lacI-S-R were used to identify and sequence the correctness of the strain, respectively. TWF067 was constructed from TWF001 by inserting the gene aspC controlled by the promoter of cysJ (PcysJ) and RBSs8 into the lacI locus in chromosome. The fragment of RBSs8-aspC was amplified by using the primers cysJ-aspC-F2/cysH-aspC-R2, and using the genomic DNA of TWF001 as a template. The promoter of cysJ was amplified by using the primers cysJ-aspC-F1/cysJ-aspC-R1, and using the genomic DNA of TWF001 as a template. These two DNA fragments were linked together by overlap PCR, using the primers cys-aspC-F/cys-aspC-R, resulting in the replacement fragment PcysJ-RBSs8-aspC. TWF068 was constructed from TWF001 by inserting the gene aspC controlled by the promoter of cysD (PcysD) and RBSs9 into the lacI locus in chromosome. The fragment of RBSs9-aspC was amplified by using the primers cysD-aspC-F2/cysH-aspC-R2, and using the genomic DNA of TWF001 as a template. The promoter of cysD was amplified by using the primers cysD-aspC-F1/cysD-aspC-R1, using the genomic DNA of TWF001 as a template. These two DNA fragments were linked together by overlap PCR, using the primers cys-aspC-F/cys-aspC-R, resulting in the replacement fragment PcysD-RBSs9-aspC.
Flask cultivation for l-threonine production in E. coli strains TWF063, TWF066, TWF067 and TWF068. a Genetic comparison of the strains constructed by dynamic regulation gene aspC. b Cell growth; c glucose consumption; dl-threonine production; e relative transcription levels of aspC. The error bars indicate the standard deviations from three independent experiments
As shown in Fig. 5a, TWF069, TWF070, TWF071, TWF072 and TWF073 were derived from TWF066 using the same method to construct TWF051, TWF052, TWF053, TWF054 and TWF055, respectively. TWF074, TWF075, TWF076 and TWF077 were derived from TWF070 using the same method to construct TWF051, TWF053, TWF054 and TWF055, respectively. TWF078, TWF079 and TWF080 were derived from TWF077 using the same method to construct TWF051, TWF053 and TWF054, respectively. TWF081 and TWF082 were derived from TWF078 using the same method to construct TWF053 and TWF054, respectively. TWF083 was derived from TWF081 using the same method to construct TWF054.
Flask cultivation for l-threonine production in E. coli strains TWF066, TWF069, TWF070, TWF071, TWF072, TWF073, TWF074, TWF075, TWF076, TWF077, TWF078, TWF079, TWF080, TWF081, TWF082 and TWF083. a Genetic comparison of the strains constructed by dynamic regulation genes arcA, cpxR, gadE, pykF and fadR. b Cell growth; cl-threonine production. The error bars indicate the standard deviations from three independent experiments
Flask fermentation and fed-batch fermentation
Escherichia coli strains were streaked from the frozen stock on LB plate and cultured at 37 °C overnight. Then, a loop of bacteria was transferred into a tube containing 5 mL LB medium and cultured at 37 °C for 5 h with 200 rpm shaking. Next, the culture was transferred into a 250-mL flask containing 30 mL LB medium (the initial OD600 of 0.05) and cultured at the same growth condition for 6 h. Finally, 5 mL culture was transferred into a 500-mL flask containing 30 mL fermentation medium (2 g/L yeast extract, 2 g/L citric acid, 25 g/L (NH4)2SO4, 7.46 g/L KH2PO4, 40 g/L glucose, 2 g/L MgSO4·7 H2O, 5 mg/L FeSO4·7H2O, 5 mg/L MnSO4·4 H2O, and 20 g/L CaCO3, pH 6.8) [50] at 37 °C with 200 rpm shaking for flask fermentation, or the whole culture was transferred into a quadruple bioreactor (Parallel-Bioreactor, China) containing 1 L fermentation medium (30 g/L glucose, 20 g/L (NH4)2SO4, 3 g/L yeast extract, 2 g/L KH2PO4, 2 g/L MgSO4·7H2O, 5 mg/L FeSO4·7H2O, 5 mg/L MnSO4·4 H2O) [6]. Temperature was maintained at 37 °C, the aeration rate at 1.5 vvm, pH was maintained automatically at 6.9 with NH4OH, and the dissolved oxygen value was maintained below 30%. Biomass was characterized by the OD600 value. One unit of OD600 corresponds to 1.7 g/L cell wet weight [6]. The amount of amino acids was determined by the 1200 series HPLC system (Agilent Technology, USA), using the orthophthalaldehyde precolumn derivatization method [51].
Acetate, pyruvate and oxaloacetate were quantified by using 1200 Series HPLC system (Agilent Technology, USA) equipped with an amines HPX-87H column (300 × 7.8 mm), and 0.005 M H2SO4 was used as a mobile phase with a flow rate of 0.5 mL/Min. The column temperature was maintained at 40 °orthophthalaldehyde and the UV absorption was determined at 210 nm.
Quantification of mRNA
The transcriptional levels of iclR, arcA, cpxR, gadE, pykF, fadR and aspC in different E. coli strains was quantified using the real-time PCR (RT-PCR). Total RNA was extracted from different E. coli cells grown at 9 h and 18 h. According to the published method [52, 53], the relative abundance of the targeted mRNAs was quantified based on the cycle threshold value, which is defined as the number of cycles required to obtain a fluorescent signal above the background and was calculated. The relative abundance of 16S rRNA was used as an internal standard. All assays were performed in triplicate.
Statistical analysis
Experimental data were expressed as means ± standard deviations. Statistical comparisons were made by one-way ANOVA to detect significant difference for l‐threonine production and glucose consumption between control and experimental groups. Spots with an adjusted p-value<0.05 were considered statistically significant.
Results
Expression regulation of arcA, cpxR, gadE, pykF and fadR to increase l-threonine production in E. coli TWF001
The thr operon controls the expression of l-threonine synthesis in E. coli by thrL [11]. ArcA represses some genes involved in TCA cycle and glyoxylate shunt [34], and l-threonine production increased in the arcA mutant TWF010 [46]. Pyruvate kinase encoded by pykF is important for cellular metabolism and glycolysis in E. coli, and the pykF mutant THRD produced more l-threonine [48]. FadR regulates the transcription of most genes involving in fatty acid degradation [28], and the mutant TWF044 in which both iclR and fadR were deleted could increase l-threonine production [6]. CpxR and GadE can activate the genes involved in the type II fatty acid synthase systems [32, 33]. Since the genes arcA, cpxR, gadE, pykF and fadR are important for l-threonine biosynthesis in E. coli, it is necessary to dynamically regulate their expression. In this study, the RBS sequence of the five genes arcA, cpxR, gadE, pykF and fadR in TWF001 was individually replaced with thrR, resulting in the stains TWF051, TWF052, TWF053, TWF054 and TWF055, respectively (Fig. 2a).
After 30 h flask fermentation, the maximum OD600 of E. coli TWF001 reached 16.67 and produced 14.75 g/L l-threonine from 40 g/L glucose (Fig. 2). Compared with TWF001, the maximum OD600 (15.88) of arcA deletion mutant TWF010 [46] decreased, but the maximum l-threonine production of TWF010 increased (Fig. 2). TWF010 consumed glucose faster than TWF001 and other five mutant strains, and used up all the glucose after 18 h; after 30 h, glucose was completely consumed in all strains (Fig. 2c).
The maximum l-threonine productions in TWF051, TWF052, TWF053, TWF054 and TWF055 were all lager than TWF001 and TWF010 (Fig. 2d). This indicates that the expression regulation of these five genes is beneficial for l-threonine biosynthesis in E. coli.
The transcriptional levels of arcA in TWF051, cpxR in TWF052, gadE in TWF053, pykF in TWF054, and fadR in TWF055 were determined (Fig. 2e). All these genes were up-regulated in the early stage of fermentation (9 h), but down-regulated in the later stage (18 h). This indicates that the dynamic regulation of the target gene worked in all five mutants and explains the higher l-threonine production in these mutants, although the degree of regulation varies with the target gene. The maximum l-threonine production (16.99 g/L) was obtained in TWF052 with a yield of 0.424 g/g glucose.
Expression regulation of iclR to increase l-threonine production in E. coli TWF001
In previous work, we have developed an l-threonine-producing strain TWF003 from E. coli TWF001 by deleting iclR. The l-threonine production in TWF003 reached 11.76 g/L, which is a 26% increase compared to TWF001 [9]. Deleting fadR [6] or arcA [46] in TWF003 could further increase l-threonine production. Since IclR regulates the expression of the aceBAK operon which could induce the glyoxylate bypass, and this could bypass prevent the quantitative loss of the entering carbon as CO2 in the Krebs cycle [54], the deletion of iclR might influence the cell growth of E. coli; therefore, dynamically regulating the expression of iclR should be a good strategy to balance the cell growth and l-threonine production in TWF001. Since IclR also regulates its own expression, we tried to change its regulatory region to regulate the expression of iclR according to the intracellular l-threonine concentration. Starting from TWF001, TWF056 was constructed by inserting thrR1 between RBSiclR and iclR, TWF057 was constructed by inserting thrR2 between PiclR and RBSiclR, TWF058 was constructed by replacing RBSiclR with thrR, and TWF059 was constructed by replacing PiclR and RBSiclR with Pthrl-thrR (Fig. 3a).
During flask fermentation, the maximum OD600 of TWF056, TWF057, TWF058 and TWF059 reached 16.12, 15.94, 16.24 and 16.34, respectively, which are higher than that of TWF001 but lower than that of TWF003 [9] (Fig. 3b). The results indicate that the autoregulation of iclR expression in these mutants did not significantly affect the cell growth.
Glucose consumption patterns are quite different in these strains. Similar levels of glucose were consumed in these strains after 9 h. After 18 h glucose was completely consumed in TWF003, TWF060, TWF061, TWF062, and TWF064. After 30 h, glucose was used up in all strains (Fig. 3c). During flask fermentation, TWF056, TWF057, TWF058 and TWF059 produced more l-threonine than TWF001 and TWF003 (Fig. 3d). The results indicate that dynamic regulation of iclR expression is better than deleting iclR to improve l-threonine biosynthesis in E. coli. Among these four strains, TWF058 produced the highest amount of l-threonine (16.95 g/L) after 18 h flask cultivation, which is 6.34% more than that of TWF003. This suggests that the dynamic regulatory iclR expression by replacing RBSiclR with thrR is a good choice for improving l-threonine production in E. coli.
To understand the difference on growth rate and l-threonine productions in the above mutant strains, the transcriptional level of iclR were determined by RT-PCR (Fig. 3e). Compared to the control TWF001, the transcriptional level of iclR in all four mutant strains increased after 9 h but decreased after 18 h. This suggests that the expression of iclR in these strains were dynamically regulated, i.e., the expression of iclR was up-regulated in the early stage of the growth but down-regulated in the late stage of the growth.
In order to optimize the dynamic regulation of iclR expression, different RBS in thrR were designed (Table 4). The mutants TWF060, TWF061, TWF062, TWF063, TWF064 and TWF065 were constructed from TWF058 by replacing RBSiclR with thrRs1, thrRs2, thrRs3, thrRs4, thrRs5, and thrRs6, respectively (Table 1 and Fig. 3a). After 30 h cultivation, the maximum OD600 of TWF060, TWF061, TWF062, TWF063, TWF064 and TWF065 reached 14.90, 13.95, 13.95, 17.62, 15.17 and 16.62, respectively; the maximum production of l-threonine in TWF060, TWF061, TWF062, TWF063, TWF064 and TWF065 reached 16.96, 15.34, 16.58, 17.81, 17.15 and 17.12, respectively. Among these mutants, TWF063is the best according to the growth rate and l-threonine productions.
The transcriptional levels of iclR in TWF060, TWF061, TWF062, TWF063, TWF064 and TWF065 were also determined by RT-PCR, using TWF001 as the control. Compared to TWF001, the transcription levels of iclR in TWF061, TWF062, TWF063, TWF064 and TWF065 were up-regulated in the early stage of the growth (9 h) but down-regulated in the late stage (18 h). This indicates again that the regulation of iclR in these mutants satisfies the growth needs in the early stage of fermentation as well as the needs for l-threonine production in the late stage of fermentation. The maximum l-threonine production (17.81 g/L) was obtained in TWF063 with a yield of 0.445 g/g glucose.
Expression regulation of aspC to improve l-threonine production in E. coli TWF063
Previously, l-threonine producing E. coli strain TWF006 was constructed from TWF004 by inserting the trc promoter in the upstream of aspC in chromosome [9]. The l-threonine production in TWF006 reached 12.47 g/L from 30 g/L glucose after 36 h flask fermentation, which is 11.6% increase compared to the control TWF004 [9]. More l-threonine was also produced when Ptac-aspC was inserted into the lacZ locus of E. coli TWF041 [6]. Since the expression regulation of aspC is related to the cell morphology and growth [55], the dynamic expression regulation of aspC is necessary to improve l-threonine production in E. coli. Therefore, the promoters of cysH, cysJ and cysD which can be activated by l-threonine [12] were fused with different RBS sequences to dynamically regulate the l-threonine production in TWF063. The fragments PcysH-RBSs7-aspC, PcysJ-RBSs8-aspC or PcysD-RBSs9-aspC were individually inserted into the lacI loci in TWF063, resulting in strains TWF066, TWF067 and TWF068, respectively (Fig. 4a).
Glucose consumption patterns are quite similar in these strains. After 30 h, glucose was used up in all strains (Fig. 4c). After 30 h flask fermentation, TWF063 produced 16.34 g/L l-threonine from 40 g/L glucose and its maximum OD600 reached 15.30 with glucose being completely consumed (Fig. 4). TWF066 grew (15.16) similar with TWF063 but produced more l-threonine (17.56 g/L) than TWF063. TWF067 and TWF068 grew slower than TWF063, and TWF067 produced less l-threonine (15.25 g/L) than TWF063, but TWF068 produced more l-threonine (17.00 g/L) than TWF063. The results indicate that the regulation of these three promoters was different, and different expression of aspC could affect the growth and l-threonine production. To understand the difference on growth and l-threonine productions in the above mutant strains, the transcriptional level of apsC was determined by RT-PCR (Fig. 4e). Compared to the control TWF063, the transcriptional level of aspC was increased in all three mutant strains during fermentation. At 9 h, the expressions of aspC in TWF066 and TWF068 were lower than that in TWF067, while at 18 h, the expression of aspC in TWF066 and TWF068 was higher than that in TWF067. Among the three mutants, the expression difference of aspC between 9 h and 18 h in TWF066 was the largest, which is consistent with the highest production of l-threonine in TWF066 This suggests that the promoter of cysH is most sensitive to l-threonine among the three promoters. The maximum l-threonine production (17.56 g/L) was obtained in TWF066 with a yield of 0.439 g/g glucose.
Expression of multiple genes to improve l-threonine production in E. coli
Since dynamic regulation of arcA, cpxR, gadE, fadR or pykF could increase l-threonine production in TWF001 (Fig. 2), these genes were also designed to be dynamically regulated in TWF066, resulting in strains TWF069, TWF070, TWF071, TWF072 and TWF073 (Fig. 5a).
After 30 h flask fermentation, TWF066 can produce 17.35 g/L l-threonine from 40 g/L glucose and its maximum OD600 reached 15.29 with glucose being completely consumed. The maximum OD600 of TWF069, TWF070, TWF071, TWF072 and TWF073 reached 15.63, 16.75, 15.46, 15.74 and 15.55, respectively. TWF069, TWF070, TWF071, TWF072 and TWF073 produced 17.94, 18.66, 17.37, 17.78 and 17.51 g/L l-threonine, respectively. Among the five mutants, TWF070 is the best based on cells growth and l-threonine yield (Fig. 5).
Mutant strains TWF074 (regulating arcA), TWF075 (regulating gadE), TWF076 (regulating pykF) and TWF077 (regulating fadR) was constructed from TWF070 (Fig. 5a). The maximum OD600 of TWF074, TWF075, TWF076 and TWF077 reached 16.56, 16.32, 15.38 and 16.34, respectively. TWF074, TWF075, TWF076 and TWF077 produced 19.61, 19.25, 19.23 and 20.36 g/L l-threonine, respectively. Among the four mutants, TWF077 produced the highest l-threonine yield.
Mutant strains TWF078 (regulating arcA), TWF079 (regulating gadE) and TWF080 (regulating pykF) was constructed from TWF077. The maximum OD600 of TWF078, TWF079 and TWF080 reached 16.41, 16.19 and 16.82, respectively (Fig. 5b). TWF078 and TWF080 produced more l-threonine (21.81 and 20.70 g/L, respectively) than TWF077.
From TWF078, mutant strains TWF081 (regulating gadE) and TWF082 (regulating pykF) was constructed. From TWF081, mutant strain TWF083 (regulating pykF) was further constructed (Fig. 5a). The maximum OD600 of TWF081, TWF082 and TWF083 reached 15.42, 16.07 and 15.68, respectively. TWF081 and TWF082 produced less l-threonine (20.94 and 21.24 g/L, respectively) than TWF078, but TWF083 produced more l-threonine (22.90 g/L) than TWF078. Among all the mutants, TWF083 produced the highest l-threonine yield.
Comparison of some key l-threonine producing E. coli strains
To compare their high l-threonine production, flask fermentations of some key mutant strains were performed (Fig. 6). Compared with the control TWF001 [56], all mutant strains produced more l-threonine, and TWF083 produced the most (Fig. 6b). The highest specific growth rate was observed around 3 h for all strains, and then the specific growth rate decreased with time (Fig. 6a). The highest specific l-threonine production rate was observed between 9 and 15 h in these strains (Fig. 6b). During 6 h to 15 h, the specific l-threonine production rate of TWF078 was higher than TWF083, but the final production of l-threonine was lower than TWF083; possibly because that the l-threonine production of TWF078 (0.774 g/L) was much lower than that of TWF083 (0.813 g/L) at 3 h. The acetate concentration in the mutant strains during the fermentation was quite different from that of TWF001, and much less acetate was accumulated in TWF083 (Fig. 6c). The pyruvate concentration in the mutant strains during the fermentation was also quite different from that of TWF001 (Fig. 6d). TWF083 and TWF078 produced more pyruvate than other strains during 6–24 h. This data indicates that in the mutant strains, especially TWF083, more carbon source was used for l-threonine production. TWF083 and TWF078 also produced more oxaloacetate, the precursor for l-threonine synthesis, than other strains during the fermentation (Fig. 6e). The intracellular NADPH content of TWF078 and TWF083 were lower than that of other strains before 18 h, suggesting that they consumed more NADPH for l-threonine biosynthesis (Fig. 6f).
Comparison of cell growth (a), l-threonine production (b), acetate concentration (c), pyruvate concentration (d), oxaloacetate concentration (e), and NADPH level (f) during flask fermentation of TWF001, TWF058, TWF063, TWF066, TWF070, TWF077, TWF078 and TWF083. The error bars indicate the standard deviations from three independent experiments
Effect of glucose concentration on l-threonine production in E. coli TWF083
TWF001 and TWF083 were grown in medium with different glucose concentrations (Fig. 7). When the initial glucose concentration in the medium increased from 30 to 50 g/L, the cell growth pattern of TWF001 changed. when grown in medium containing 30, 40, and 50 g/L initial glucose, glucose in TWF001 was completely consumed after 15, 24 and 30 h, and the maximum OD600 of TWF001 reached 13.80, 14.76, and 13.96, respectively (Fig. 7a–c). The cell growth and glucose consumption patterns of TWF083 were quite different from TWF001 (Fig. 7). When the initial glucose was 30 g/L, the glucose was depleted after 18 h, and the highest OD600 reached 15.44. When the initial glucose was 40 g/L, the glucose was depleted after 21 h, and the highest OD600 reached 17.44 When the initial glucose was 50 g/L, the glucose was depleted after 21 h, and the highest OD600 reached 16.36).
Comparison for l-threonine production in E. coli strains TWF001 and TWF083 under different glucose addition. a 30 g/L glucose; b 40 g/L glucose; c 50 g/L glucose; d 2 * 30 g/L glucose, the first was added at the beginning and the second when the glucose was used up completely, respectively; e 2 * 40 g/L glucose, the first was added at the beginning and the second at 18 h, respectively; f 50 and 30 g/L glucose, the first was added at the beginning and the second at 21 h, respectively. The error bars indicate the standard deviations from three independent experiments
l-Threonine productions in TWF001 and TWF083 grown in medium containing with different concentrations of glucose are also shown in Fig. 7. Under the same growth conditions, TWF083 always produced more l-threonine than TWF001. The highest l-threonine production was obtained in either TWF001 or TWF083 when grown in medium containing 40 g/L initial glucose. When grown in medium with 30 g/L initial glucose for 18 h, TWF083 produced 18.76 g/L l-threonine, while TWF001 produced 15.86 g/L l-threonine. When grown in medium with 40 g/L initial glucose for 21 h, TWF083 produced 26.50 g/L l-threonine, and the conversion rate of glucose to l-threonine reached 0.66 g/g, while TWF001 produced 17.74 g/L l-threonine, and the conversion rate of glucose to l-threonine reached 0.44 g/g. When grown in medium with 50 g/L initial glucose for 27 h, TWF083 produced 26.93 g/L l-threonine, while TWF001 produced 16.58 g/L l-threonine. The results suggest that TWF083 could efficiently consume glucose to produce l-threonine.
When TWF083 grown in the medium containing 40 g/L glucose for 21 h, glucose was depleted, and l-threonine production reached the highest (Fig. 7b). Therefore, the fermentation of TWF083 was conducted in medium with a second addition of glucose, using TWF001 as a control (Fig. 7). TWF001 and TWF083 cells were grown in medium with 30 g/L initial glucose, and 30 g/L glucose was added when the initial glucose was depleted at 15 and 18 h, respectively. The highest l-threonine production in TWF001 and TWF083 reached 19.67 and 23.11 g/L, respectively (Fig. 7d). TWF001 and TWF083 cells were grown in medium with 40 g/L initial glucose, and 40 g/L glucose was added when the initial glucose was depleted at 18 h. The highest l-threonine production in TWF001 and TWF083 reached 18.90 and 29.73 g/L, respectively (Fig. 7e). TWF001 and TWF083 cells were grown in medium with 50 g/L initial glucose, and 30 g/L glucose was added when the initial glucose was depleted at 21 h. The highest l-threonine production in TWF001 and TWF083 reached 18.72 and 7.33 g/L, respectively (Fig. 7f). Compared with the control TWF001, TWF083 grown better and produced more l-threonine under the same growth condition.
Fed‐batch fermentation of TWF083 was performed in 2.4-L quadruple fermenters with a working volume of 1 L, using TWF001 as a control (Fig. 8). TWF001 could produce 82.9 g/L l‐threonine in 48 h, leading to a yield of 0.357 g/g glucose and a volumetric productivity of 1.73 g/L/h. TWF083 produced 116.62 g/L l‐threonine with a yield of 0.486 g/g glucose and productivity of 2.43 g/L/h. Both biomass accumulation and glucose consumption of TWF083 were faster than TWF001 in the fermenter, suggesting that the dynamic regulation expression of multiple key genes in TWF083 functions well in large-scale fermentation.
Discussion
In this study, the thrL regulatory region was used as dynamic regulation element to control the expression of some key genes responsive to l-threonine. The gene arcA in TCA cycle, the genes fadR, cpxR and gadE in fatty acid synthesis and the gene pykF in aerobic respiration were repressed using the thrR dynamic regulation. The RBSs with different strength in thrR were designed to optimize the dynamic regulation of the gene iclR in the glyoxylate shunt. The gene aspC following same strength RBSs and different threonine-activating promoters, PcysH, PcysJ and PcysD, were inserted into the chromosome to enhance its expression. Finally, all regulations were integrated into one strain, and the resulting strain TWF083 could produce 26.50 g l-threonine from 40 g glucose, which is 49.4% higher than the control TWF001, and the conversion rate reached 0.66 g/g (Fig. 7b). In recent years, many studies have focused on designing genetic circuits to realize dynamic regulation of metabolic flux for producing intermediates [57] such as lycopene [58], myoinositol [59] and l-lysine [60]. A series of threonine-responsive sensors were derived by transforming thrL leader regulatory elements, which were in the negative correlation with l-threonine concentration and has different sensitivity [61]. However, there was no report on the application of thrL regulation in l-threonine production.
The modification of glyoxylate shunt has been used to overcome acetate overflow and improve the production of acetyl-CoA-derived chemicals by deleting iclR in E. coli [9, 22,23,24, 26]. In this study, the strain TWF056, TWF057, TWF058, TWF059, TWF060, TWF061, TWF062, TWF063, TWF064 and TWF065 were constructed by inserting the different part of attenuator region of thr operon, thrL, into the different native regulatory regions in front of iclR. All these strains could produce more l-threonine than the control TWF001 after 30 h fermentation. TWF063 produced 11.7% more l-threonine than the iclR deletion mutant TWF003 [9], and its growth was also improved (Fig. 3).
The aspartate aminotransferase encoded by aspC draws the carbon flux from TCA cycle to the biosynthetic pathway of l-aspartate family. Deletion of aspC led to generation of small cells with slow growth, while overexpression of aspC exerted the opposite effect [55]. Using a switch to make the glycolytic fluxes towards TCA cycle in the early stage, and to make the carbon flux be redirected into l-threonine synthetic pathway by inducing the expression of aspC, fdh, gdhA and pntAB in the production stage could increase l-threonine production [62]. The gene aspC following different threonine-enhancing promoters were inserted into the chromosome of TWF063, resulting in the strains TWF066, TWF067 and TWF068. Because of the high expression of aspC in early stage, the growth of TWF067 and TWF068 was inhibited and less l-threonine was produced, but the lower expression of aspC at the early stage and the higher expression at the later stage made the growth of TWF066 unaffected, but the l-threonine production increased by 7% (Fig. 4).
The global regulation factor arcA deletion has been used in E. coli to activate the TCA cycle and aerobic respiration [46, 63]. The global regulation factor fadR deletion has been used to increase the acetyl-CoA supply by enhancing the fatty acid degradation [6]. The role of cpxR and gadE in fatty acid synthesis is to activate fabA and fabZ [32, 33], which are not regulated by fadR, but there was no report on the accumulation of substances by knocking out cpxR and gadE. In pykF knockout mutant, less acetate and more l-threonine was produced [47, 48]. With the dynamic regulation, the nature regulatory regions of the above 5 genes were replaced by thrL leader regulatory elements in TWF001, resulting in the strains TWF051, TWF052, TWF053, TWF054 and TWF055. All these mutants could produce more l-threonine than the control TWF001 after 30 h fermentation. TWF066 was used as a starting strain, step by step, to carry out multiple knockout test on the 5 genes to find the optimal strain. The results showed that the genetic engineering through the dynamic regulation had little effect on the metabolism of the strain in the early stage. After several rounds of modifications, the growth of strain TWF083 was similar to that of original strain TWF001 but the l-threonine production increased by 49.4%.
Several l-threonine producing E. coli strains have been constructed. An l-methionine auxotroph strain KY10935 obtained by a classical selection method without the use of directed genetic-engineering modifications could produce 100 g/L l-threonine after 77 h cultivation [65]. TH28C/pBRThrABCR3 constructed from W3110 by systems metabolic engineering and in silico flux response analysis could produce threonine with a high yield of 0.393 g/g glucose and 82.4 g/L threonine by fed-batch culture [8]. KCCM 10353 could produce 112 g/L l-threonine with a high yield of 0.452 g/g glucose after 77 h cultivation [66]. EC125 could produce 105.3 g/L l-threonine with a yield of 0.405 g/g glucose after 48 h two-stage feeding cultivation [67]. THRDΔpykF produced less acetate (52%) and produced 112.57 g/L l-threonine with a yield of 0.376 g/g glucose after 40 h cultivation [48]. TRFC derived by repeated compound mutagenesis (DES plus UV) from E. coli K12 could use sucrose as the carbon source, it could produce 124.57 g/L l-threonine after 40 h cultivation, using the combined feeding strategy of pseudo-exponential feeding and glucose-stat feeding resulted in high cell density [68]. THPE5 uses a switch to make the glycolytic fluxes towards TCA cycle in the early stage, leading to the improved glucose utilization and growth performance; in the production stage the carbon flux is redirected into l-threonine synthetic pathway via a synthetic genetic circuit by inducing the expression of aspC, fdh, gdhA and pntAB to increase l-threonine production, resulting in 70.5 g/L l-threonine with a yield of 0.404 g/g glucose after 40 h cultivation [62]. JLTHR could produce 127.3 g/L l-threonine with a glucose conversion rate of 58.12% after adding betaine hydrochloride in the medium [4]. There are a few l-threonine producing strains were developed from the same origin to TWF083. TWF044 could enhance glyoxylate shunt pathway and produce 103.89 g/L l-threonine with a yield of 0.450 g/g glucose after 48 h fed-batch fermentation [6]. TWF006/pFW01-thrA*BC-asd could produce only 15.85 g/L l-threonine but showed a high yield of 0.528 g/g glucose after 36 h shake flask cultivation [9]. TWF018 could produce 25.1 g/L l-threonine with a high yield of 0.628 g/g glucose after 36 h shake flask cultivation [46]. WMZ016/pFW01-thrA*BC-rhtC derived from MG1655 by modifying the glucose specific phosphotransferase system could produce 17.98 g/L l-threonine with a yield of 0.346 g/g glucose after 36 h shake flask cultivation [69]. When TWF083 was cultured in shake flask, it produced 16.50 g/L l-threonine with a yield of 0.662 g/g glucose after 30 h, which is better than other strains derived from the same origin. After 48 h fed-batch fermentation, TWF083 could produce 116.62 g/L l-threonine with a yield of 0.486 g/g glucose.
NADPH is essential for the catalytic activity of glutamate dehydrogenase in E. coli [70]. It also plays a key role in l‐threonine biosynthesis due to the NADPH‐dependent β‐aspartate semialdehyde dehydrogenase and homoserine dehydrogenase [71, 72]. In E. coli, NADPH is mostly produced in the pentose phosphate pathway by glucose-6-phosphate dehydrogenase encoded by the gene zwf, which catalyzes the oxidation of glucose 6-phosphate to 6-phosphoglucono-δ-lactone; 6-phosphogluconate dehydrogenase encoded by the genes gnd could also produce NADPH through the oxidative decarboxylation of 6-phosphogluconate to ribulose 5-phosphate [73]. Several approaches to increase the NADPH yield through metabolic engineering of genes involved in the pentose phosphate pathway have been reported [74,75,76,77,78,79,80]. Therefore, it is worth studying the influence of NADPH on l-threonine production in TWF083.
In this study, a range of approaches have been proposed for the construction of l-threonine producing strain using dynamic regulation. The successful application of threonine-responsive elements to improve l-threonine production in this work provides further opportunities for synthetic biology and metabolic engineering in E. coli.
Availability of data and materials
All data generated or analyzed during this study are included in the manuscript.
References
Dong X, Quinn PJ, Wang X. Microbial metabolic engineering for l-threonine production. Subcell Biochem. 2012;64:283–302.
Lee M-H, Lee H-W, Park J-H, Ahn J-O, Jung J-K, Hwang Y-I. Improved l-threonine production of Escherichia coli mutant by optimization of culture conditions. J Biosci Bioeng. 2006;101(2):127–30.
Dong X, Quinn PJ, Wang X. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for the production of l-threonine. Biotechnol Adv. 2011;29(1):11–23.
Su Y, Guo QQ, Wang S, Zhang X, Wang J. Effects of betaine supplementation on l-threonine fed-batch fermentation by Escherichia coli. Bioprocess Biosyst Eng. 2018;41(10):1509–18.
Zhang X, Yan J, Yu L, Zhang G, Zhang Y, Chen N, Wen T. Construction of recombinant plasmids containing threonine operon and their effects on l-threonine accumulation. Acta Microbiol Sin. 2009;49:591.
Yang J, Fang Y, Wang J, Wang C, Zhao L, Wang X. Deletion of regulator-encoding genes fadR, fabR and iclR to increase l-threonine production in Escherichia coli. Appl Microbiol Biotechnol. 2019;103:4549–64.
Livshits VA, Zakataeva NP, Aleshin VV, Vitushkina MV. Identification and characterization of the new gene rhtA involved in threonine and homoserine efflux in Escherichia coli. Res Microbiol. 2003;154:123–35.
Lee KH, Park JH, Kim TY, Kim HU, Lee SY. Systems metabolic engineering of Escherichia coli for l-threonine production. Mol Syst Biol. 2007;3:149.
Zhao H, Fang Y, Wang X, Zhao L, Wang J, Li Y. Increasing l-threonine production in Escherichia coli by engineering the glyoxylate shunt and the L-threonine biosynthesis pathway. Appl Microbiol Biotechnol. 2018;102:5505–18.
Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43:867–79.
Lynn SP, Burton WS, Donohue TJ, Gould RM, Gumport RI, Gardner JF. Specificity of the attenuation response of the threonine operon of Escherichia coli is determined by the threonine and isoleucine codons in the leader transcript. J Mol Biol. 1987;194:59–69.
Liu YN, Li Q, Zheng P, Zhang Z, Liu Y, Sun C, Cao G, Zhou W, Wang X, Zhang D, et al. Developing a high-throughput screening method for threonine overproduction based on an artificial promoter. Microb Cell Fact. 2015;14:121.
Mohsin M, Ahmad A. Genetically-encoded nanosensor for quantitative monitoring of methionine in bacterial and yeast cells. Biosens Bioelectron. 2014;59:358–64.
Mustafi N, Frunzke J. Development and application of a single-cell biosensor for the detection of l-methionine and branched-chain amino acids. Metab Eng. 2012;14:449–57.
Binder S, Schendzielorz G, Stabler N, Krumbach K, Hoffmann K, Bott M, Eggeling L. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol. 2012;13:R40.
Hoffmann K, Grunberger A, Lausberg F, Bott M, Eggeling L. Visualization of imbalances in sulfur assimilation and synthesis of sulfur-containing amino acids at the single-cell Level. Appl Environ Microbiol. 2013;79:6730–6.
Garcia JR, Cha HJ, Rao G, Marten MR, Bentley WE. Microbialnar-GFP cell sensors reveal oxygen limitations in highly agitated and aerated laboratory-scale fermentors. Microb Cell Fact. 2009;8:6–6.
Binder S, Siedler S, Marienhagen J, Bott M, Eggeling L. Recombineering in Corynebacterium glutamicum combined with optical nanosensors: a general strategy for fast producer strain generation. Nucleic Acids Res. 2013;41:6360.
Schendzielorz G, Dippong M, Grünberger A, Kohlheyer D, Eggeling L. Taking control over control: use of product sensing in single cells to remove flux control at key enzymes in biosynthesis pathways. ACS Synth Biol. 2014;3:21.
Lee JH, Wendisch VF. Production of amino acids-Genetic and metabolic engineering approaches. Bioresour Technol. 2017;245:1575–87.
Yamamoto K, Ishihama A. Two different modes of transcription repression of the Escherichia coli acetate operon by IclR. Mol Microbiol. 2003;47:183–94.
Liu M, Ding Y, Chen H, Zhao Z, Liu H, Xian M, Zhao G. Improving the production of acetyl-CoA-derived chemicals in Escherichia coli BL21(DE3) through iclR and arcA deletion. BMC Microbiol. 2017;17:10.
Li Y, Huang B, Wu H, Li Z, Ye Q, Zhang YP, Li Y, Huang B, Wu H, Li Z. Production of succinate from acetate by metabolically engineered Escherichia coli. ACS Synth Biol. 2016;5:1299.
Zhang T, Wang Z, Deng L, Tan T, Wang F, Yan Y. Pull-in urea cycle for the production of fumaric acid in Escherichia coli. Appl Microbiol Biotechnol. 2015;99:5033–44.
Woo SC, In KD, Sol C, Jae Won J, Yup LS. Metabolic engineering of Escherichia coli for the production of fumaric acid. Biotechnol Bioeng. 2013;110:2025–34.
Ning Y, Wu X, Zhang C, Xu Q, Chen N, Xie X. Pathway construction and metabolic engineering for fermentative production of ectoine in Escherichia coli. Metab Eng. 2016;36:10–8.
Fengming L, Yu C, Robert L, Kilho L, Yingjin Y, Xiaoxia Nina L. Improving fatty acid availability for bio-hydrocarbon production in Escherichia coli by metabolic engineering. PLoS ONE. 2013;8:e78595.
Campbell JW, Morgan-Kiss RM, Cronan JE. A new Escherichia coli metabolic competency: growth on fatty acids by a novel anaerobic β-oxidation pathway. Mol Microbiol. 2010;47:793–805.
Campbell JW, Cronan JE. Escherichia coli fadR positively regulates transcription of the fabB fatty acid biosynthetic gene. J Bacteriol. 2001;183:5982–90.
My L, Ghandour Achkar N, Viala JP, Bouveret E, de Boer P. Reassessment of the genetic regulation of fatty acid synthesis in Escherichia coli: global positive control by the dual functional regulator FadR. J Bacteriol. 2015;197:1862–72.
My L, Rekoske B, Lemke JJ, Viala JP, Gourse RL, Bouveret E. Transcription of the Escherichia coli fatty acid synthesis operon fabHDG is directly activated by FadR andinhibited by ppGpp. J Bacteriol. 2013;195:3784–95.
Hommais F. GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology. 2004;150:61.
Dartigalongue C, Missiakas D, Raina S. Characterization of the Escherichia coli σE Regulon. J Biol Chem. 2001;276:20866–75.
Iuchi S, Lin EC. arcA (dye), arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci USA. 1988;85:1888–92.
Park SJ, McCabe J, Turna J, Gunsalus RP. Regulation of the citrate synthase (gltA) gene of Escherichia coli in response to anaerobiosis and carbon supply: role of the arcA gene product. J Bacteriol. 1994;176:5086–92.
Cunningham L, Gruer MJ, Guest JR. Transcriptional regulation of the aconitase genes (acnA and acnB) of Escherichia coli. Microbiology. 1997;143:3795–805.
Chao G, Shen J, Tseng CP, Park SJ, Gunsalus RP. Aerobic regulation of isocitrate dehydrogenase gene (icd) expression in Escherichia coli by the arcA and fnr gene products. J Bacteriol. 1997;179:4299–304.
Park SJ, Chao G, Gunsalus RP. Aerobic regulation of the sucABCD genes of Escherichia coli, which encode alpha-ketoglutarate dehydrogenase and succinyl coenzyme A synthetase: roles of ArcA, Fnr, and the upstream sdhCDAB promoter. J Bacteriol. 1997;179:4138–42.
Spencer ME, Guest JR. Transcription analysis of the sucAB, aceEF and lpd genes of Escherichia coli. Mol Gen Genet. 1985;200:145.
Park SJ. Regulation of succinate dehydrogenase (sdhCDAB) operon expression in Escherichia coli in response to and anaerobiosis: role of ArcA and Fnr. Mol Microbiol. 2010;15:473–82.
Park SJ, Gunsalus RP. Oxygen, iron, carbon, and superoxide control of the fumarase fumA and fumC genes of Escherichia coli: role of the arcA, fnr, and soxR gene products. J Bacteriol. 1995;177:6255–62.
Tseng CP. Regulation of fumarase (fumB) gene expression in Escherichia coli in response to oxygen, iron and heme availability: role of the arcA, fur and hemA gene products. FEMS Microbiol Lett. 2010;157:67–72.
Park SJ, Cotter PA, Gunsalus RP. Regulation of malate dehydrogenase (mdh) gene expression in Escherichia coli in response to oxygen, carbon, and heme availability. J Bacteriol. 1995;177:6652.
Pan B, Unnikrishnan I, Laporte DC. The binding site of the IclR repressor protein overlaps the promoter of aceBAK. J Bacteriol. 1996;178:3982–4.
Quail MA, Haydon DJ, Guest JR. The pdhR-aceEF-lpd operon of Escherichia coli expresses the pyruvate dehydrogenase complex. Mol Microbiol. 2010;12:95–104.
Ding Z, Fang Y, Zhu L, Wang J, Wang X. Deletion of arcA, iclR, and tdcC in Escherichia coli to improve l-threonine production. Biotechnol Appl Biochem. 2019;66:794–807.
Siddiquee KA, Arauzo-Bravo MJ, Shimizu K. Effect of a pyruvate kinase (pykF-gene) knockout mutation on the control of gene expression and metabolic fluxes in Escherichia coli. FEMS Microbiol Lett. 2004;235:25–33.
Xie XX, Liang Y, Liu H, Liu Y, Xu Q, Zhang C, Chen N. Modification of glycolysis and its effect on the production of l-threonine in Escherichia coli. J Ind Microbiol Biotechnol. 2014;41:1007–15.
Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol. 2015;81:2506–14.
Lee JH, Sung BH, Kim MS, Blattner FR, Yoon BH, Kim JH, Kim SC. Metabolic engineering of a reduced-genome strain of Escherichia coli for l-threonine production. Microb Cell Fact. 2009;8:2.
Kőrös Á, Varga Z, Molnár-Perl I. Simultaneous analysis of amino acids and amines as their o-phthalaldehyde-ethanethiol-9-fluorenylmethyl chloroformate derivatives in cheese by high-performance liquid chromatography. J Chromatogr A. 2008;1203:146–52.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8.
Nolden L, Farwick M, Krämer R, Burkovski A. Glutamine synthetases of Corynebacterium glutamicum: transcriptional control and regulation of activity. FEMS Microbiol Lett. 2001;201:91–8.
Gui L, Sunnarborg A, Pan B, Laporte DC. Autoregulation of iclR, the gene encoding the repressor of the glyoxylate bypass operon. J Bacteriol. 1996;178:321–4.
Liu F, Qimuge JH, Hao J, Yan H, Bach T, Fan L, Morigen. AspC-mediated aspartate metabolism coordinates the Escherichia coli cell cycle. PLoS ONE. 2014;9:e92229.
Zhao L, Zhang H, Wang X, Han G, Ma W, Hu X, Li Y. Transcriptomic analysis of an l-threonine-producing Escherichia coli TWF001. Biotechnol Appl Biochem. 2020. https://doi.org/10.1002/bab.1890.
Dahl RH, Zhang F, Alonso-Gutierrez J, Baidoo E, Batth TS, Redding-Johanson AM, Petzold CJ, Mukhopadhyay A, Lee TS, Adams PD, Keasling JD. Engineering dynamic pathway regulation using stress-response promoters. Nat Biotechnol. 2013;31:1039.
Farmer W, Liao J. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat Biotechnol. 2000;18:533–7.
Gupta A, Reizman IMB, Reisch CR, Prather KLJ. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat Biotechnol. 2017;35:273.
Zhou LB, Zeng AP. Exploring lysine riboswitch for metabolic flux control and improvement of l-lysine synthesis in Corynebacterium glutamicum. ACS Synth Biol. 2015;4:729–34.
Liu Z, Zheng Y, Chen T, Ma H. Transform thrL leading regulatory elements for different threonine sensor. Biotechnology. 2016;26:372–9.
Liu J, Li H, Xiong H, Xie X, Chen N, Zhao G, Caiyin Q, Zhu H, Qiao J. Two-stage carbon distribution and cofactor generation for improving l-threonine production of Escherichia coli. Biotechnol Bioeng. 2018;116:110–20.
Yao P, Cui Y, Yu S, Du Y, Feng J, Wu Q, Zhu D. Efficient biosynthesis of (R)- or (S)-2-hydroxybutyrate froml-threonine through a synthetic biology approach. Adv Synth Catal. 2016;358:2923–8.
Espah Borujeni A, Cetnar D, Farasat I, Smith A, Lundgren N, Salis HM. Precise quantification of translation inhibition by mRNA structures that overlap with the ribosomal footprint in N-terminal coding sequences. Nucleic Acids Res. 2017;45:5437–48.
Okamoto K, Kino K, Ikeda M. Hyperproduction of l-threonine by an Escherichia coli mutant with impaired l-threonine uptake. Biosci Biotechnol Biochem. 1997;61:1877–82.
Yuzbashev TV, Vybornaya TV, Larina AS, Gvilava IT, Voyushina NE, Mokrova SS, Yuzbasheva EY, Manukhov IV, Sineoky SP, Debabov VG. Directed modification of Escherichia coli metabolism for the design of threonine-producing strains. Appl Biochem Micro. 2013;49:723–42.
Liu S, Liang Y, Liu Q, Tao T, Lai S, Chen N, Wen T. Development of a two-stage feeding strategy based on the kind and level of feeding nutrients for improving fed-batch production of l-threonine by Escherichia coli. Appl Microbiol Biotechnol. 2013;97:573–83.
Wang J, Cheng LK, Chen N. High-level production of l-threonine by recombinant Escherichia coli with combined feeding strategies. Biotechnol Biotechnol Equip. 2014;28:495–501.
Zhu L, Fang Y, Ding Z, Zhang S, Wang X. Developing an l-threonine-producing strain from wild-type Escherichia coli by modifying the glucose uptake, glyoxylate shunt, and l-threonine biosynthetic pathway. Biotechnol Appl Biochem. 2019;66:962–76.
Wang P, Yang X, Lin B, Huang J, Tao Y. Cofactor self-sufficient whole-cell biocatalysts for the production of 2-phenylethanol. Metab Eng. 2017;44:143–9.
Bartek T, Blombach B, Zonnchen E, Makus P, Lang S, Eikmanns BJ, Oldiges M. Importance of NADPH supply for improved l-valine formation in Corynebacterium glutamicum. Biotechnol Prog. 2010;26:361–71.
Takeno S, Murata R, Kobayashi R, Mitsuhashi S, Ikeda M. Engineering of Corynebacterium glutamicum with an NADPH-generating glycolytic pathway for l-lysine production. Appl Environ Microbiol. 2010;76:7154–60.
Stephanopoulos GN, Aristidou AA, Nielsen J. Chapter 2—Review of cellular metabolism. San Diego: Academic Press; 1998. p. 21–79.
Lim SJ, Jung YM, Shin HD, Lee YH. Amplification of the NADPH-related genes zwf and gnd for the oddball biosynthesis of PHB in an E. coli transformant harboring a cloned phbCAB operon. J Biosci Bioeng. 2002;93:543–9.
Lee JN, Shin HD, Lee YH. Metabolic engineering of pentose phosphate pathway in Ralstonia eutropha for enhanced biosynthesis of poly-β-hydroxybutyrate. Biotechnol Prog. 2003;19:1444–9.
Poulsen BR, Nøhr J, Douthwaite S, Hansen LV, Iversen JJL, Visser J, Ruijter GJG. Increased NADPH concentration obtained by metabolic engineering of the pentose phosphate pathway in Aspergillus niger. FEBS J. 2005;272:1313–25.
Li Y, Zhang D, Cai N, Han C, Mao Q, Wang T, Zhou Q, Chen N, Xie X. Betaine supplementation improved l-threonine fermentation of Escherichia coli THRD by upregulating zwf (glucose-6-phosphate dehydrogenase) expression. Electron J Biotechnol. 2019;39:67–73.
Fabrizio C, Hess TA, Sylvia H, Wang T, Thomas S, Uwe S. Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microbiol Lett. 2001;204(2):247–52.
Lee HC, Kim JS, Jang W, Kim SY. High NADPH/NADP(+) ratio improves thymidine production by a metabolically engineered Escherichia coli strain. J Bacteriol. 2010;149:24–32.
Siedler S, Bringer S, Bott M. Increased NADPH availability in Escherichia coli: improvement of the product per glucose ratio in reductive whole-cell biotransformation. Appl Microbiol Biotechnol. 2011;92:929–37.
Funding
This study was supported by the National Key Research and Development Program of China (2018YFA0900300) and the National First-class Discipline Program of Light Industry Technology and Engineering (LITE2018-10).
Author information
Authors and Affiliations
Contributions
LZ, YL and XW designed the experiments. JY, YF, LZ, ZD, CW, WM and XH participated in the discussion about the experiments. LZ performed experiments and drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhao, L., Lu, Y., Yang, J. et al. Expression regulation of multiple key genes to improve l-threonine in Escherichia coli. Microb Cell Fact 19, 46 (2020). https://doi.org/10.1186/s12934-020-01312-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12934-020-01312-5