Abstract
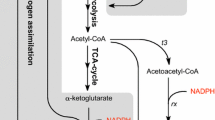
A basic requirement for the efficiency of reductive whole-cell biotransformations is the reducing capacity of the host. Here, the pentose phosphate pathway (PPP) was applied for NADPH regeneration with glucose as the electron-donating co-substrate using Escherichia coli as host. Reduction of the prochiral β-keto ester methyl acetoacetate to the chiral hydroxy ester (R)-methyl 3-hydroxybutyrate (MHB) served as a model reaction, catalyzed by an R-specific alcohol dehydrogenase. The main focus was maximization of the reduced product per glucose yield of this pathway-coupled cofactor regeneration with resting cells. With a strain lacking the phosphoglucose isomerase, the yield of the reference strain was increased from 2.44 to 3.78 mol MHB/mol glucose. Even higher yields were obtained with strains lacking either phosphofructokinase I (4.79 mol MHB/mol glucose) or phosphofructokinase I and II (5.46 mol MHB/mol glucose). These results persuasively demonstrate the potential of NADPH generation by the PPP in whole-cell biotransformations.



Similar content being viewed by others
References
Akinterinwa O, Cirino PC (2011) Anaerobic obligatory xylitol production in Escherichia coli strains devoid of native fermentation pathways. Appl Environ Microbiol 77:706–709
Blank LM, Ebert BE, Bühler B, Schmid A (2008) Metabolic capacity estimation of Escherichia coli as a platform for redox biocatalysis: constraint-based modeling and experimental verification. Biotechnol Bioeng 100:1050–1065
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Bühler B, Park JB, Blank LM, Schmid A (2008) NADH availability limits asymmetric biocatalytic epoxidation in a growing recombinant Escherichia coli strain. Appl Environ Microbiol 74:1436–1446
Canonaco F, Hess TA, Heri S, Wang T, Szyperski T, Sauer U (2001) Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microbiol Lett 204:247–252
Chin JW, Cirino PC (2011) Improved NADPH supply for xylitol production by engineered Escherichia coli with glycolytic mutations. Biotechnol Prog 27:333–341
Chin JW, Khankal R, Monroe CA, Maranas CD, Cirino PC (2009) Analysis of NADPH supply during xylitol production by engineered Escherichia coli. Biotechnol Bioeng 102:209–220
Eguchi T, Kuge Y, Inoue K, Yoshikawa N, Mochida K, Uwajima T (1992) NADPH regeneration by glucose dehydrogenase from Gluconobacter scleroides for l-leucovorin synthesis. Biosci Biotechnol Biochem 56:701–703
Ernst M, Kaup B, Müller M, Bringer-Meyer S, Sahm H (2005) Enantioselective reduction of carbonyl compounds by whole-cell biotransformation, combining a formate dehydrogenase and a (R)-specific alcohol dehydrogenase. Appl Microbiol Biotechnol 66:629–634
Fasan R, Crook NC, Peters MW, Meinhold P, Buelter T, Landwehr M, Cirino PC, Arnold FH (2010) Improved product-per-glucose yields in P450-dependent propane biotransformations using engineered Escherichia coli. Biotechnol Bioeng 108:500–510
Fraenkel DG, Levisohn SR (1967) Glucose and gluconate metabolism in an Escherichia coli mutant lacking phosphoglucose isomerase. J Bacteriol 93:1571–1578
Fraenkel DG, Kotlarz D, Buc H (1973) Two fructose 6-phosphate kinase activities in Escherichia coli. J Biol Chem 248:4865–4866
Fuhrer T, Sauer U (2009) Different biochemical mechanisms ensure network-wide balancing of reducing equivalents in microbial metabolism. J Bacteriol 191:2112–2121
Fuhrer T, Fischer E, Sauer U (2005) Experimental identification and quantification of glucose metabolism in seven bacterial species. J Bacteriol 187:1581–1590
Grose JH, Joss L, Velick SF, Roth JR (2006) Evidence that feedback inhibition of NAD kinase controls responses to oxidative stress. Proc Natl Acad Sci U S A 103:7601–7606
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580
Hanahan D, Jessee J, Bloom FR (1991) Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol 204:63–113
Heuser F, Schroer K, Lütz S, Bringer-Meyer S, Sahm H (2007) Enhancement of the NAD(P)(H) pool in Escherichia coli for biotransformation. Eng Life Sci 7:343–353
Holm AK, Blank LM, Oldiges M, Schmid A, Solem C, Jensen PR, Vemuri GN (2010) Metabolic and transcriptional response to cofactor perturbations in Escherichia coli. J Biol Chem 285:17498–17506
Hua Q, Yang C, Baba T, Mori H, Shimizu K (2003) Responses of the central metabolism in Escherichia coli to phosphoglucose isomerase and glucose-6-phosphate dehydrogenase knockouts. J Bacteriol 185:7053–7067
Kaup B, Bringer-Meyer S, Sahm H (2004) Metabolic engineering of Escherichia coli: construction of an efficient biocatalyst for D-mannitol formation in a whole-cell biotransformation. Appl Microbiol Biotechnol 64:333–339
Kaup B, Bringer-Meyer S, Sahm H (2005) D-Mannitol formation from D-glucose in a whole-cell biotransformation with recombinant Escherichia coli. Appl Microbiol Biotechnol 99:397–403
Kruger NJ, von Schaewen A (2003) The oxidative pentose phosphate pathway: structure and organisation. Curr Opin Plant Biol 6:236–246
Lee HC, Kim JS, Jang W, Kim SY (2010) High NADPH/NADP+ ratio improves thymidine production by a metabolically engineered Escherichia coli strain. J Biotechnol 149:24–32
Lee JN, Shin HD, Lee YH (2003) Metabolic engineering of pentose phosphate pathway in Ralstonia eutropha for enhanced biosynthesis of poly-beta-hydroxybutyrate. Biotechnol Prog 19:1444–1449
Liljeblad A, Kallinen A, Kanerva LT (2009) Biocatalysis in the preparation of the statin side chain. Curr Org Synth 6:362–379
Lim SJ, Jung YM, Shin HD, Lee YH (2002) Amplification of the NADPH-related genes zwf and gnd for the oddball biosynthesis of PHB in an E. coli transformant harboring a cloned phbCAB operon. J Biosci Bioeng 93:543–549
Link AJ, Phillips D, Church GM (1997) Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol 179:6228–6237
Lundquist R, Olivera BM (1971) Pyridine nucleotide metabolism in Escherichia coli. I. Exponential growth. J Biol Chem 246:1107–1116
Matsumura H, Miyachi S (1980) Cycling assay for nicotinamide adenine dinucleotides. Meth Enzymol 69:465
Miller, J (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, pp. 352–355
Morita T, El-Kazzaz W, Tanaka Y, Inada T, Aiba H (2003) Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J Biol Chem 278:15608–15614
Nor'Aini AR, Shirai Y, Hassan MA, Shimizu K (2006) Investigation on the metabolic regulation of pgi gene knockout Escherichia coli by enzyme activities and intracellular metabolite concentrations. Malays J Microbiol 2:24–31
Panke S, Wubbolts M (2005) Advances in biocatalytic synthesis of pharmaceutical intermediates. Curr Opin Chem Biol 9:188–194
Park J, Gupta RS (2008) Adenosine kinase and ribokinase—the RK family of proteins. Cell Mol Life Sci 65:2875–2896
Poulsen RB, Nøhr J, Douthwaite S, Hansen LV, Iversen JJL, Visser J, Ruijter GJG (2005) Increased NADPH concentration obtained by metabolic engineering of the pentose phosphate pathway in Aspergillus niger. FEBS J 272:1313–1325
Roehl RA, Vinopal RT (1976) Lack of glucose phosphotransferase function in phosphofructokinase mutants of Escherichia coli. J Bacteriol 126:852–860
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Santamaría B, Estevez AM, Martinez-Costa OH, Aragon JJ (2002) Creation of an allosteric phosphofructokinase starting with a nonallosteric enzyme. The case of Dictyostelium discoideum phosphofructokinase. J Biol Chem 277:1210–1216
Sauer U, Canonaco F, Heri S, Perrenoud A, Fischer E (2004) The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J Biol Chem 279:6613–6619
Stephanopoulos GN, Aristidou AA, Nielsen J (1998) Review of cellular metabolism. In: Stephanopoulos GN, Aristidou AA, Nielsen J (eds) Metabolic engineering: principles and methodologies. Academic, San Diego, pp 21–79
Vinopal RT, Fraenkel DG (1975) PfkB and pfkC loci of Escherichia coli. J Bacteriol 122:1153–1161
Vinopal RT, Clifton D, Fraenkel DG (1975) PfkA locus of Escherichia coli. J Bacteriol 122:1162–1171
Walton AZ, Stewart JD (2004) Understanding and improving NADPH-dependent reactions by nongrowing Escherichia coli cells. Biotechnol Prog 20:403–411
Zerez CR, Moul DE, Gomez EG, Lopez VM, Andreoli AJ (1987) Negative modulation of Escherichia coli NAD kinase by NADPH and NADH. J Bacteriol 169:184–188
Zhang W, O'Connor K, Wang DI, Li Z (2009) Bioreduction with efficient recycling of NADPH by coupled permeabilized microorganisms. Appl Environ Microbiol 75:687–694
Acknowledgements
This work was funded by the BIO.NRW Cluster Biotechnology North Rhine-Westphalia, Germany (support code: W0805wb001b).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1A
Intracellular concentrations of NADPH and NADP+ and ratios of [NADPH]/[NADP+] in E. coli reference strain and deletion mutants at three time points during biotransformation. All strains expressed the plasmid-encoded alcohol dehydrogenase gene from Lactobacillus brevis. Samples taken at the time point zero did not yet contain the biotransformation substrate MAA. Results were derived from at least two independent experiments (PDF 16 kb)
Table S1B
Intracellular concentrations of NADH and NAD+ and ratios of [NADH]/[NAD+] in E. coli reference strain and deletion mutants at three time points during biotransformation. All strains expressed the plasmid-encoded alcohol dehydrogenase gene from Lactobacillus brevis. Samples taken at the time point zero did not yet contain the biotransformation substrate MAA. Results were derived from at least two independent experiments (PDF 16 kb)
Fig. S1
Correlation of the [NADPH]/[NADP+] ratios before MAA addition to the biotransformation test mixtures (t 0) and the biotransformation yield (mole MHB per mole glucose) (PDF 19 kb)
Rights and permissions
About this article
Cite this article
Siedler, S., Bringer, S. & Bott, M. Increased NADPH availability in Escherichia coli: improvement of the product per glucose ratio in reductive whole-cell biotransformation. Appl Microbiol Biotechnol 92, 929–937 (2011). https://doi.org/10.1007/s00253-011-3374-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3374-4