Abstract
Background
The characterization of the molecular determinants of metal resistance has potential biotechnological application in biosensing and bioremediation. In this context, the bacterium Thermus thermophilus HB27 is a metal tolerant thermophile containing a set of genes involved in arsenic resistance which, differently from other microbes, are not organized into a single operon. They encode the proteins: arsenate reductase, TtArsC, arsenic efflux membrane transporter, TtArsX, and transcriptional repressor, TtSmtB.
Results
In this work we show that the arsenic efflux protein TtArsX and the arsenic responsive transcriptional repressor TtSmtB are required to provide resistance to cadmium. We analyzed the sensitivity to Cd(II) of mutants lacking TtArsX, finding that they are more sensitive to this metal than the wild type strain. In addition, using promoter probe reporter plasmids, we show that the transcription of TtarsX is also stimulated by the presence of Cd(II) in a TtSmtB-dependent way. Actually, a regulatory circuit composed of TtSmtB and a reporter gene expressed from the TtarsX promoter responds to variation in Cd(II), As(III) and As(V) concentrations.
Conclusions
Our results demonstrate that the system composed by TtSmtB and TtArsX is responsible for both the arsenic and cadmium resistance in T. thermophilus. The data also support the use of T. thermophilus as a suitable chassis for the design and development of As-Cd biosensors.
Similar content being viewed by others
Background
Toxic metals and metalloids such as cadmium (Cd) and arsenic (As) are widespread environmental contaminants that pose risks to human health [1]. Microorganisms are endowed with multiple molecular mechanisms to handle exposure to these toxic compounds. In general, microbial resistance is achieved through three main mechanisms: transformation of the metals through reduction to a different oxidation state, efflux outside the cell by transporters, and/or sequestration/biosorption [2]. Common reduction mechanisms include for example the conversion of Hg2+ to Hg0, AsO43− to AsO33−. Facilitated efflux transporters fall into two wide, functionally and evolutionary distinct membrane protein families, the P-type ATPases and the Major Facilitator Superfamily (MFS) of transporters [3,4,5]. P-type ATPases use ATP hydrolysis to transport ions across cellular membranes and are composed of three conserved domains: (1) the transmembrane (TM) helix bundle, allowing substrate translocation; (2) the soluble ATP binding domain (ATPBD) that contains the transiently phosphorylated Asp residue; (3) the soluble actuator domain (AD) [6]. Over the years, on the basis of sequence similarity and overall architecture, they have been divided into different classes: those belonging to P1B-type are capable to drive the efflux out of cells of both essential transition metal ions (e.g., Zn2+, Cu+, and Co2+) and toxic metal ions (e.g., Ag+, Cd2+, Pb2+) contributing to their homeostasis maintenance. A recent study on a huge number of P1B-type ATPase sequences combined with available biochemical data classifies them into seven distinct subfamilies (1B-1 1B-7) on the basis of conserved motifs in TM4, TM5 and TM6, but the molecular basis of metal ion specificity remains unclear [7]. All metal efflux transporters characterized so far are tightly controlled at transcriptional level by metalloregulatory proteins which bind DNA sequences and dissociate following metal binding, thus ensuring derepression of genes encoding efflux proteins [8]. Several regulatory systems dedicated to metal/metalloid sensing have also been characterized; for instance, transcription factors of the ArsR/SmtB family are small dimeric proteins with a winged helix-turn-helix DNA binding domain controlling gene expression in response to divalent metals (e.g. zinc, cadmium) as well as metalloids (e.g. arsenic and antimonite) through an allosteric switching mechanism [9]. The exploration of life in extreme environments has led to the isolation of many thermophilic microorganisms occupying diverse extreme habitats like hot hydrothermal fluids containing high concentrations of toxic metals. For this reason they are able to cope with toxic metals, which are more soluble at high temperatures, or even to use them for their metabolism [10, 11] and are currently exploited in some bioprocesses such as biomining and bioremediation [12]. A detailed understanding of the molecular mechanisms responsible for resistance to toxic metals is also crucial for engineering organisms to develop sensitive biosensors for the detection of chemicals in the environment and to enhance bioremediation strategies [13, 14].
A significant number of whole-cell arsenic and/or cadmium biosensors has been already described in literature and is based on the realization of reporter systems containing regulatory cis-acting sequences interacting with a transcriptional repressor belonging to the ArsR/SmtB family [15, 16]. Biosensors are not intended to fully replace chemical methods but have the advantage of lower fabrication cost and higher stability and can offer on-site monitoring of even trace levels of targeted compounds in comparison with non-portable analytical methodologies [17]. To date, the major challenges in biosensor development regard the screening or modification of efficient regulator protein/promoter pair for increased sensitivity and specificity [18, 19] as well as biosensor stability over time and simultaneous monitoring of multiple environmental parameters [20].
Thermus thermophilus HB27 is a thermophilic aerobic Gram-negative bacterium capable of growing in the presence of arsenic concentrations that are lethal for other microorganisms [21]. In recent studies we demonstrated that the arsenic resistance system is not clustered in a single ars operon as in other organisms, but the genes are spread in the genome: TTC1502 encoding a cytoplasmic arsenate reductase (TtArsC) able to reduce arsenate to arsenite, TTC0354, encoding a P1B-type membrane ATPase responsible for the efflux (herein named TtArsX) and TTC0353 encoding a repressor (TtSmtB) sensitive to both As(V) and As(III) [22,23,24]. TtSmtB is a member of the ArsR/SmtB family, sharing 50% identity with the well characterized SmtB of Synechococcus PCC7942 [25]. It is a dimeric protein containing three Cys residues in a reduced state and a conserved metal binding box presumably involved in As(V) and As(III) binding. The protein can bind to a consensus regulatory sequence located upstream of TtarsX preferentially in an un-metallated state and in vivo TtSmtB regulates TtarsX transcription upon arsenic interaction through a derepression mechanism [24].
In the present study, we evaluated the contribution of TtSmtB and TtArsX in cadmium detoxification using a combination of genetic and physiological approaches. The results obtained support that in T. thermophilus the mechanism employed for survival to cadmium and arsenic exposure is promiscuous, suggesting that the evolution of shared/common defense mechanisms represents an adaptation strategy to cope with toxic metals at high temperatures whereas keeping a reduced genome. In addition, to analyze the cadmium/arsenic response of TtarsX promoter in dependency on varying TtSmtB concentration, we settled up different β-gal reporter systems with the final goal of evaluating the utilization of thermophilic molecular components and thermostable chassis cells in biosensing applications.
Methods
Culture conditions and determination of minimum inhibitory concentration
Strains, genotypes and sources are summarized in Additional file 1: Table S1.
T. thermophilus HB27 wild type strain, T. thermophilus ΔsmtB::kat and TTC0354::pK18 mutants were grown aerobically at 70 °C in TM medium as described [24].
Minimum inhibitory concentration (MIC) was determined as the lowest concentration of cadmium that completely inhibited the visible growth of the strains after overnight incubation as indicated by the lack of turbidity. Basically, exponentially growing cultures of T. thermophilus HB27, T. thermophilus ΔsmtB::kat and TTC0354::pK18 were diluted to 0.08 OD600 nm in 24 well plates containing increasing concentrations of CdCl2 ranging from 0 mM to 5 mM and incubated at 70 °C or 60 °C for 18 h; depending on the strain tolerance, the concentration interval was narrowed in consecutive experiments; the MIC endpoint was considered as the lowest Cd(II) concentration at which there was a difference between grown and start culture lower than 0.01OD600 nm. The values reported are the average of two independent determinations.
In silico analysis
BlastP analyses were performed using the Blastall (v.2.2.25) program. The predicted presence and number of TMs, in the full length TtArsX, was determined using the TMHMM 2.0 online (http://www.cbs.dtu.dk/services/TMHMM) and the TM-pred online servers (http://www.ch.embnet.org/software/TMPRED_form.html) [26, 27]. Conserved motifs in TM helices of TtArsX were determined by manual inspection referring to those identified by Smith et al. [7]. The presence of Metal Binding Domain(s) (MBD) was determined by manually looking at a CXXC motif in the protein sequence and fold prediction performed at (http://pfam.sanger.ac.uk).
Models of the MBD were generated using I-TASSER web server [28] (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) using as input the first 91 amino acids of TtArsX. The model was selected according to its similarity to available crystal structures of MBD of P1B-type ATPases from other species [6].
The docking calculations were obtained using Hex Protein Docking server [29] with TtArsX MBD and As(III) or Cd(II); 100 rigid body docking solutions were generated and the 10 best refined by energy minimization. The proposed model for the metal docked into the MBD is the structure with the smallest distance between the metal and cysteine [2.61 Å for As(III) or 2.35 Å for Cd(II)], after an energy minimization step.
Bioreporter constructions
The regulatory region upstream of TtarsX (TtarsXp) previously named TTC0354p, was amplified by PCR using the primer pairs new 0354pr fwEcoRI and R0354NdeI, respectively (Additional file 1: Table S2); the region extends from − 73 to + 1 from the transcription start site [24]. The primers introduced EcoRI and NdeI restriction sites, so that the amplified fragment could be cloned in the adapted pMHbgaA plasmid [30]. The new vector was named pMHTtarsXpbgaA.
To obtain the plasmid pMHTtarsXpbgaA-nqoTtsmtB, the pnqo-TtsmtB gene cassette, where TtsmtB is under the control of the nqo promoter, was cloned into pMHTtarsXpbgaA vector; in T. thermophilus, the nqo promoter drives the expression of the operon encoding the major respiratory complex I during aerobic growth [31]. In particular, pET28/TtsmtB was digested with NdeI HindIII [24] and cloned into the corresponding sites of pMKpnqo-bgaA [31] giving pMKpnqo-TtsmtB; afterwards, the plasmid was digested with XbaI HindIII and the gel purified pnqo-TtsmtB cassette subjected to a fill-in reaction and cloned into the filled-in HindIII site of pMHTtarsXpbgaA.
pMHTtarsXpbgaA-nqoTtSmtB was used to transform T. thermophilus HB27 and TTC0354::pK18 mutants in the conditions described [24]. The pMHPnorbgaA vector was also used to transform the same strains and used as negative control [32]. Cells were then incubated for 24–48 h at 60 °C on TM plates containing hygromycin (100 μg/mL). All the plasmids used in this study are described in Additional file 1: Table S3.
β-galactosidase assays
For measuring reporter β-galactosidase activity, the growing transformants were diluted to 0.1 OD600 nm in TM medium in the presence of hygromycin (100 μg/ml), treated with different concentrations of NaAsO2, KH2AsO4 and CdCl2 (Sigma) as source of As(III), As(V) and Cd(II) respectively, and grown at 60 °C for 16 h. β-galactosidase activity assays were carried out on permeabilized cells in 96-well microplates at 70 °C with a Synergy H4 microplate reader (BioTeK) as described by Miller [33].
Miller units (U) were calculated by the equation:
where: OD420 = OD of the chromogenic product, OD550 = OD of the cellular debris, t = time of reaction, V = volume of used cells and OD600 = OD of the cell culture. β-galactosidase activity of T. thermophilus negative control (transformed with pMHPnorbgaA vector) was subtracted from that of the samples. The activity reported is the average of two or three independent experiments each made in triplicate. The error bars indicate the standard deviation of the average values. Miller Units expressed as a percentage, were calculated assuming that the Miller Units value of not treated cells (control) was 100%. Statistical analysis was performed using a Student’s t test; significant differences are indicated as: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Purification of TtSmtB
Recombinant TtSmtB was purified to homogeneity using the procedure already described, consisting of thermo-precipitation of the E. coli BL21-Codon Plus (DE3) RIL/TtSmtB cell extract followed by HiTrap Heparin chromatography. The histidine tag was removed from purified TtSmtB by thrombin digestion (Sigma). The purified protein was stored in aliquots at − 20 °C [24].
Electrophoretic mobility shift assay (EMSA)
To determine if cadmium was ligand of TtSmtB, electrophoretic mobility shift assays (EMSA) were performed. The TtarsX promoter region was amplified by PCR using specific primer pair: 0354footprint fw and 0354footprint rv, (Additional file 1: Table S2). EMSA reactions were set up as described [34], using 5 µM of proteins pre-incubated or not with Cd(II) at molar ratio of 1:20 and 1:50 (considering TtSmtB as a dimer).
Results and discussion
Domain organization and subfamily classification of TtArsX
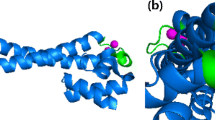
Our recent work showed that TTC0354, herein named TtArsX, is a membrane metal-transporter involved in arsenic detoxification; in fact, a mutant strain in which this gene had been knocked out, was about 15-fold more sensitive to both As(III) and As(V) treatment than the wild type [24]. In order to analyse the putative role of this protein in the detoxification of metals different from As(III)/As(V), in particular Cd(II), at first some in silico studies were performed aimed at identifying divergence or not in conserved motifs. TM helix prediction tools suggested that TtArsX is composed of six TM helices containing all the conserved signatures of the characterized P1B-2-subclass members which generally display a dual role in Zn2+ transport and toxic metal ion detoxification, especially Cd(II) [7, 35]. They are: SXP and CPC motifs in TM4, a T(X)5QN(X)7K motif in TM5 and a DXG(X)7N in TM6 (Fig. 1a in bold). A previous work had already shown that TtArsX is a P1B-type ATPase stimulated in vitro by Zn2+/Cd2+ cations but with an unclear role in their tolerance in vivo [23]. The overall topology of TtArsX (Fig. 1b) obtained integrating results from a 3D model of the protein [24] also helped to identify a soluble MBD containing the CXXC motif that, as indicated by docking analyses, could be responsible for both Cd(II) and As(III) recognition inside the cell (Fig. 1c).
a Primary structure of TtArsX. Aminoacids that are predicted to be in β-turns are colored green, those in α-helices are red and those in turns are colored black. Aminoacids in orange are included in TM helices. The residues conserved in P-type ATPases are shown in bold, those which are signatures of the P1b-2 type subfamily are underlined. The CXXC motif at the N-terminus is boxed. b Schematic topology of TtArsX (adapted from [7]). MBD: soluble N-terminal metal-binding domain (hexagon); AD: actuator domain (ellipse), ATPBD: ATP binding domain. Invariant motifs in ATPBD and AD domains of P-type ATPases are shown in bold. Residues indicated in the TM helices 4,5,6 are those conserved in P1b-2 type subfamily. c 3D model of TtArsX MBD (corresponding to residue 1–91 from the intact protein) interacting with As(III), (colored blue, left) and Cd(II) (magenta, right)
Taken together, the results of in silico analysis assign TtArsX to the well characterized P1B-2-subfamily, show the presence of a soluble heavy metal associated domain (Pfam: PF 00403) and suggest a wider metal ion specificity than that previously known.
Contribution of TtArsX and TtSmtB to the Cd(II) tolerance mechanism
To analyse the role of TtArsX and the transcriptional repressor TtSmtB [24] in resistance to cadmium, we used different physiological and genetic approaches.
First, we compared the growth of T. thermophilus HB27 and two mutants defective in TtArsX or TtSmtB (T. thermophilus TTC0354::pK18 and T. thermophilus ΔsmtB::kat, respectively) in the presence of different Cd(II) concentrations, and determined the MIC values which are reported in Table 1. T. thermophilus TTC0354::pK18 revealed a 15-fold increase in cadmium sensitivity in comparison to the wild type, thus showing a role of TtArsX in Cd(II) resistance; on the other hand, T. thermophilus ΔsmtB::kat, showed a 3-fold increase in cadmium sensitivity as compared to wild type HB27. One possible explanation for this unexpected result is a polar effect of the TtsmtB::kat mutation: in the mutant strain the kanamycin resistance gene is in counter sense; therefore, the basal levels of TtArsX could be lower than in the wild type, making the cells more sensitive.
In addition, the comparison of MIC values with those previously reported for As(V) and As(III) showed that T. thermophilus HB27 is almost 14-fold more sensitive to Cd(II) than to arsenic (Table 1).
To analyse in vivo whether TtarsX promoter had Cd(II) responsive activity, the regulatory region was cloned in a promoter probe vector upstream of the bgaA gene, encoding a thermostable β-galactosidase [30]. The plasmid pMHTtarsXpbgaA (Fig. 2a) was transformed into T. thermophilus HB27 and the β-gal activity was measured also upon Cd(II) treatment at 10, 20 and 100 µM (see “Methods”). The results in Fig. 2b show that transcription from TtarsX promoter in the reporter system is activated by Cd(II) by twofold at 20 µM (633 ± 84 Miller Units, MU) in comparison to values in untreated cells (393 ± 40 MU). At 100 µM Cd(II) transcription is also increased in comparison to the control, but at lower levels; the reduced activity could be the consequence of partial toxicity, since reduced growth rates were observed under these conditions (Additional file 1: Figure S1).
Cd(II) dependent activity of TtarsX promoter with transcriptional bgaA fusions. a Schematic representation of the reporter system used to transform T. thermophilus HB27 and TTC0354::pK18. b β-gal activity expressed in % MU of T. thermophilus HB27-pMHTtarsXpbgaA treated or not with 10, 20 and 100 µM of CdCl2. Statistical analysis was performed using the Student’s t test; significant differences are indicated as: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
To check whether TtSmtB was the transcriptional regulator of TtArsX in the presence of Cd(II), we performed in vivo and in vitro experiments. The comparison of β-gal activities in T. thermophilus HB27-pMHTtarsXpbgaA and T. thermophilus ΔsmtB::kat-pMHTtarsXpbgaA grown in the absence of metals (393 ± 40 and 887 ± 138 MU, respectively), is consistent with the in vivo role of TtSmtB as a repressor. Moreover, EMSA assays were carried out with recombinant TtSmtB on the TtarsX promoter after preincubation with or without Cd(II). As shown in Fig. 3, our results confirm that the protein interacts with TtarsXp and indicate that, upon binding with Cd(II), binding of TtSmtB to the promoter is hampered. TtSmtB DNA-binding behavior upon interaction with Cd(II) is similar to that already observed with As(III) and As(V) [24], suggesting that metal ions could take contacts with the same protein motif. Moreover, these results strongly support the role of TtSmtB in Cd(II) mediated TtarsX transcription where it works as repressor.
Altogether, these results indicate that TtSmtB regulates cadmium tolerance by controlling at transcriptional level the metal efflux gene, adopting a derepression mechanism similar to that employed for arsenic detoxification [24].
Bioreporter construction and characterization
To evaluate the potential of the pair composed by TtSmtB and TtarsX responsive promoter as components of a bioreporter system for toxic metal detection, we aimed at characterizing their cadmium and arsenic sensing potential in engineered T. thermophilus cells. Hence, we generated a new reporter plasmid, pMHTtarsXpbgaA-nqoTtSmtB, to measure the transcription of the reporter gene from the TtarsX promoter in a context in which the TtsmtB gene was expressed constitutively from the nqo promoter (Fig. 4a). We expected that an increase in the intracellular concentration of the transcription factor allowed a more efficient repression of the system and, as a consequence, an activity of the reporter gene mainly depending on metal concentration. As shown in Fig. 4b, the system responds to increasing concentrations of Cd(II) almost in a linear form in a wide range, but with a slope too flat to measure any concentration accurately. As we suspected that this was due to the presence of TtArsX actively extruding Cd(II), we assayed the bioreporter in T. thermophilus TTC0354::pK18 defective mutant. As expected (Fig. 4c), the increase in bioreporter activity was higher in the mutant strain than in the parental one; in fact at 10 µM Cd(II) a twofold induction was measured in the first, whereas no significant induction occurs in the wild type; furthermore, the mutant strain was able to detect Cd(II) at 5 µM, a concentration value that is fourfold lower than that required to get a similar signal in the parental strain. However, the tradeoff of this system is that in the mutant strain the response decreases above 20 µM Cd(II). Therefore, it could be envisioned to use both host systems to cope with a wider range of Cd(II) concentrations. The increase in the sensitivity of a biosensor using strains with mutations in the arsenic efflux pump in place of the wild type has been also reported for P. putida [36].
Cd(II) dependent bioreporter response. a Schematic representation of the recombinant vector used to transform T. thermophilus HB27 and TTC0354::pK18. b β-gal activity expressed in % MU of T. thermophilus HB27-pMHTtarsXpbgaA-nqoTtSmtB treated or not with 10, 20 and 100 µM of CdCl2. c β-gal activity expressed in % MU of T. thermophilus TTC0354::pK18-pMHTtarsXpbgaA-nqoTtSmtB treated or not with 10, 20 and 100 µM of cadmium. Statistical analysis was performed using a Student’s t test; significant differences are indicated as: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
As previous functional studies showed that TtSmtB and TtArsX are part of the arsenic resistance system, we evaluated the bioreporter system for its capability to detect arsenate and arsenite in T. thermophilus TTC0354::pK18. Figure 5 shows that the first significant reporter signal was obtained at 20 µM As(V) and 10 µM As(III), demonstrating that inactivation of Cd/As efflux pump translates in higher response sensitivities to arsenic too. As also observed for Cd(II), the reporter activity increases to a maximum level [385 ± 22 MU for As(V) and 563 ± 65 MU for As(III)] after which it decreases likely because of the metal toxicity. Overall, this study underlines the advantage of using: (i) an extremophilic microorganism more resistant to stress conditions for the development of biosensor for in situ monitoring of contaminated sites [37]; (ii) a bioreporter system where the interaction between the regulator and its target promoter has been analyzed and defined [38]; (iii) a strain genetically modified more sensitive to detect metals [36]. In addition, these results encourage to improve the newly developed cell-based system for the realization of a robust biosensor for multiple metal detection [39]. In this context, a system to be used as a screening tool to detect metals in environmental samples could significantly help to find metal polluted areas, or to monitor in situ cleanup during bioremediation.
As(V) and As(III) dependent bioreporter response. a β-gal activity expressed in % MU of TTC0354::pK18-pMHTtarsXpbgaA-nqoTtSmtB treated or not with 10, 20, 30, 50, 100, 200 and 300 µM of As(V). b β-gal activity expressed in % MU. of T. thermophilus TTC0354::pK18-pMHTtarsXpbgaA-nqoTtSmtB treated or not with 10, 20, 30, 50, 100, 200 and 300 µM of As(III). Statistical analysis was performed using a Student’s t test; significant differences are indicated as: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Conclusions
This work reports for the first time the identification of a molecular mechanism responsible for cadmium tolerance of the thermophilic bacterium T. thermophilus, taking advantage of the utilization of suitable genetic tools. Interestingly, the system includes part of the machinery (TtArsX and TtSmtB, membrane efflux ATPase and ArsR/SmtB transcriptional repressor, respectively) used to cope with arsenic; to the best of our knowledge this is the first functional characterization in a bacterium of a common/promiscuous mechanism to defend from both arsenic and cadmium, giving new venues for the understanding of the metal response evolution and the adaptation of environmental thermophilic microorganisms to deal with high concentrations of metals under enhanced solubilization conditions [12]. Notably, from an evolutionary point of view it can be speculated that in the absence of an ars operon, cells may have evolved arsenic resistance from preexisting metal detoxification systems. It is also plausible that promiscuous detoxification systems have developed according to the hypothesis that in the genomes of thermophilic microorganisms the genetic information is condensed. Moving to biotechnologically relevant applications in the improvement of biosensor field, this study outlines the importance of a detailed characterization of the molecular components (intrinsic promoter activity, repressor/promoter and repressor/metal(s) binding affinities) and points to T. thermophilus as suitable chassis cell for design and development of robust metal biosensors. In this context, advantageous modifications can be programmed to increase biosensor sensitivity, selectivity and/or ability to detect metal mixtures.
References
Jarup L, Akesson A. Current status of cadmium as an environmental health problem. Toxicol Appl Pharmacol. 2009;238:201–8.
Das S, Dash HR, Chakraborty J. Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Appl Microbiol Biotechnol. 2016;100:2967–84.
Wong K, Ma J, Rothnie A, Biggin PC, Kerr ID. Towards understanding promiscuity in multidrug efflux pumps. Trends Biochem Sci. 2014;39:8–16.
Prasad R, Rawal MK. Efflux pump proteins in antifungal resistance. Front Pharmacol. 2014;5:202.
Paul S, Moye-Rowley WS. Multidrug resistance in fungi: regulation of transporter-encoding gene expression. Front Physiol. 2014;5:143.
Smith AT, Barupala D, Stemmler TL, Rosenzweig AC. A new metal binding domain involved in cadmium, cobalt and zinc transport. Nat Chem Biol. 2015;11:678.
Smith AT, Smith KP, Rosenzweig AC. Diversity of the metal-transporting P 1B-type ATPases. J Biol Inorg Chem. 2014;19:947–60.
Chandrangsu P, Rensing C, Helmann JD. Metal homeostasis and resistance in bacteria. Nat Rev Microbiol. 2017;15:338.
Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev. 2009;109:4644–81.
Pedone E, Bartolucci S, Fiorentino G. Sensing and adapting to environmental stress: the archaeal tactic. Front Biosci. 2004;9:2909–26.
Bartolucci S, Contursi P, Fiorentino G, Limauro D, Pedone E. Responding to toxic compounds: a genomic and functional overview of Archaea. Front Biosci. 2013;18:165–89.
Ranawat P, Rawat S. Metal-tolerant thermophiles: metals as electron donors and acceptors, toxicity, tolerance and industrial applications. Environ Sci Pollut Res 2018;25:4105–33.
Fiorentino G, Ronca R, Bartolucci S. A novel E. coli biosensor for detecting aromatic aldehydes based on a responsive inducible archaeal promoter fused to the green fluorescent protein. Appl Microbiol Biotechnol. 2009;82:67–77.
Politi J, Spadavecchia J, Fiorentino G, Antonucci I, De Stefano L. Arsenate reductase from Thermus thermophilus conjugated to polyethylene glycol-stabilized gold nanospheres allow trace sensing and speciation of arsenic ions. J R Soc Interface. 2016;13:20160629.
Choe S-I, Gravelat FN, Al Abdallah Q, Lee MJ, Gibbs BF, Sheppard DC. Role of Aspergillus niger acrA in arsenic resistance and its use as the basis for an arsenic biosensor. Appl Environ Microbiol. 2012;78:3855–63.
Ravikumar Y, prabhu Nadarajan S, Lee CS, Yun H. Engineering an FMN-based iLOV protein for the detection of arsenic ions. Anal Biochem. 2017;525:38–43.
PrÚvÚral S, Brutesco C, Descamps EC, Escoffier C, Pignol D, Ginet N, Garcia D. A bioluminescent arsenite biosensor designed for inline water analyzer. Environ Sci Pollut Res. 2017;24:25–32.
Peng Z, Yan Y, Xu Y, Takeo M, Yu H, Zhao Z, Zhan Y, Zhang W, Lin M, Chen M. Improvement of an E. coli bioreporter for monitoring trace amounts of phenol by deletion of the inducible σ54-dependent promoter. Biotech Lett. 2010;32:1265–70.
Behzadian F, Barjeste H, Hosseinkhani S, Zarei A. Construction and characterization of Escherichia coli whole-cell biosensors for toluene and related compounds. Curr Microbiol. 2011;62:690–6.
Chen J, Rosen BP. Biosensors for inorganic and organic arsenicals. Biosensors. 2014;4:494–512.
Del Giudice I, Limauro D, Pedone E, Bartolucci S, Fiorentino G. A novel arsenate reductase from the bacterium Thermus thermophilus HB27: its role in arsenic detoxification. Biochim Biophys Acta. 2013;1834:2071–9.
Politi J, Spadavecchia J, Fiorentino G, Antonucci I, Casale S, De Stefano L. Interaction of Thermus thermophilus ArsC enzyme and gold nanoparticles naked-eye assays speciation between As(III) and As(V). Nanotechnology. 2015;26:435703.
Schurig-Briccio LA, Gennis RB. Characterization of the PIB-Type ATPases present in Thermus thermophilus. J Bacteriol. 2012;194:4107–13.
Antonucci I, Gallo G, Limauro D, Contursi P, Ribeiro AL, Blesa A, Berenguer J, Bartolucci S, Fiorentino G. An ArsR/SmtB family member regulates arsenic resistance genes unusually arranged in Thermus thermophilus HB27. Microb Biotechnol. 2017;10:1690–701.
VanZile ML, Cosper NJ, Scott RA, Giedroc DP. The zinc metalloregulatory protein Synechococcus PCC7942 SmtB binds a single zinc ion per monomer with high affinity in a tetrahedral coordination geometry. Biochemistry. 2000;39:11818–29.
Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes1. J Mol Biol. 2001;305:567–80.
Hofmann K. TMbase-A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166.
Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008;9:40.
Macindoe G, Mavridis L, Venkatraman V, Devignes M-D, Ritchie DW. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res. 2010;38:W445–9.
Cava F, Zafra O, Da Costa MS, Berenguer J. The role of the nitrate respiration element of Thermus thermophilus in the control and activity of the denitrification apparatus. Environ Microbiol. 2008;10:522–33.
Cava F, Laptenko O, Borukhov S, Chahlafi Z, Blas-Galindo E, Gómez-Puertas P, Berenguer J. Control of the respiratory metabolism of Thermus thermophilus by the nitrate respiration conjugative element NCE. Mol Microbiol. 2007;64:630–46.
Alvarez L, Bricio C, Gómez MJ, Berenguer J. Lateral transfer of the denitrification pathway genes among thermus thermophilus strains. Appl Environ Microbiol. 2011;77:1352–8.
Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory: Cold Spring Harbor; 1972.
Fiorentino G, Del Giudice I, Bartolucci S, Durante L, Martino L, Del Vecchio P. Identification and physicochemical characterization of BldR2 from Sulfolobus solfataricus, a novel archaeal member of the MarR transcription factor family. Biochemistry. 2011;50:6607–21.
Smith AT, Ross MO, Hoffman BM, Rosenzweig AC. Metal selectivity of a Cd-, Co-, and Zn-transporting P1B-type ATPase. Biochemistry. 2016;56:85–95.
Fernandez M, Morel B, Ramos JL, Krell T. Paralogous regulators ArsR1 and ArsR2 of Pseudomonas putida KT2440 as a basis for arsenic biosensor development. Appl Environ Microbiol. 2016;82:4133–44.
Kim HJ, Jeong H, Lee SJ. Synthetic biology for microbial heavy metal biosensors. Anal Bioanal Chem. 2018;410:1191–203.
Arruda LM, Monteiro LM, Silva-Rocha R. The Chromobacterium violaceum ArsR arsenite repressor exerts tighter control on its cognate promoter than the Escherichia coli system. Front Microbiol. 1851;2016:7.
Amaro F, Turkewitz AP, Martin-Gonzalez A, Gutierrez JC. Whole-cell biosensors for detection of heavy metal ions in environmental samples based on metallothionein promoters from Tetrahymena thermophila. Microb Biotechnol. 2011;4:513–22.
Authors’ contributions
IA, GG and ALR performed experiments. DL, PC, AB, JB, SB and GF supervised the project. IA and GF drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors thank Dr. Emilia Pedone of the CNR Institute of Biostructures and Bioimages in Napoli for helpful discussions during the preparation of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This research was carried out in the frame of the Project “Immobilization of ENzymes on hydrophobin-functionalized NAnomaterials” funded by the University of Napoli Federico II.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional file
12934_2018_918_MOESM1_ESM.docx
Additional file 1: Table S1. Strains used in this work classified according to their genotype. Table S2. Oligonucleotides used in this work. Table S3. Plasmids used in this work classified according to their features. Figure S1. Growth curves of T. thermophilus HB27 transformed with the vector pMHTtarsXpbgaA in the absence (circle) and presence of 100 μM Cd(II) (triangle).
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Antonucci, I., Gallo, G., Limauro, D. et al. Characterization of a promiscuous cadmium and arsenic resistance mechanism in Thermus thermophilus HB27 and potential application of a novel bioreporter system. Microb Cell Fact 17, 78 (2018). https://doi.org/10.1186/s12934-018-0918-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12934-018-0918-7