Abstract
Background
Cryptosporidium and Enterocytozoon bieneusi are two important pathogens with zoonotic potential that cause enteric infections in a wide range of hosts, including humans. Both are transmitted from animals to humans by direct contact or through contaminated equipment. Bears are frequently found in Chinese zoos as ornamental animals as well as farmed as commercial animals, and are therefore in close contact with zoo- or farm-keepers, but the prevalence and zoonotic potential of Cryptosporidium and E. bieneusi in bears is poorly understood. In this study, we aimed to provide data on the occurrence and genetic diversity of Cryptosporidium and E. bieneusi in Asiatic black bears from Heilongjiang and Fujian, China. From May 2015 to December 2017, 218 fresh fecal specimens were collected from captive Asiatic black bears in Heilongjiang (n = 36) and Fujian (n = 182), China. Cryptosporidium and E. bieneusi were examined by PCR amplification of the partial small subunit of ribosomal DNA (SSU rDNA) and the internal transcribed spacer (ITS) region of rDNA, respectively. C. andersoni-positive isolates were subtyped through PCR analysis of the four minisatellite/microsatellite (MS1, MS2, MS3 and MS16) loci.
Results
The overall prevalence of Cryptosporidium and E. bieneusi were 2.4% (4/218) and 6.4% (14/218), respectively, with 2.8% (1/36) and 22.2% (8/36) in the Heilongjiang Province, and 1.6% (3/182) and 3.3% (6/182) in the Fujian Province. Sequence analysis confirmed the presence of Cryptosporidium species: C. andersoni (n = 3) and a genotype termed Cryptosporidium rat genotype IV (n = 1). All three identified C. andersoni belonged to the MLST subtype A4, A4, A4, A1. Two known E. bieneusi genotypes D (n = 4) and SC02 (n = 10) were identified, both of which belong to zoonotic Group 1.
Conclusions
This is the first report of C. andersoni and Cryptosporidium rat genotype IV in bears. The discovery of the zoonotic potential of E. bieneusi genotype D in bears highlights its significant zoonotic potential and potential threat to human health.
Similar content being viewed by others
Background
Cryptosporidium and Enterocytozoon bieneusi are two important pathogens with zoonotic potential that cause diarrhea in humans and a variety of animal species [1, 2]. For humans, both are responsible for self-limiting diarrhea in immune competent individuals, and severe life-threatening diarrhea in patients with immune deficiency [3, 4]. Cryptosporidium oocysts and E. bieneusi spores, the infective stage, are ubiquitous in the environment. Both humans and animals can be infected through direct contact with an infected host, or through indirect ingestion of oocyst-contaminated water or food [2, 3]. Current epidemiological data of Cryptosporidium and E. bieneusi have raised public health concerns about their zoonotic nature in addition to water- and food-borne transmission [2, 3]. Tracing the sources of contamination and elucidating the transmission routes of Cryptosporidium and E. bieneusi are important steps in adequately controlling human infections.
The use of PCR-based molecular tools for the characterization of Cryptosporidium and E. bieneusi have improved our understanding of their epidemiology, providing information on the host distribution of various species/genotypes and transmission routes/sources [4, 5]. To date, 39 species of Cryptosporidium and more than 70 genotypes having been described [5, 6]. Amongst them, 21 Cryptosporidium species/genotypes have been identified in humans, of which C. hominis and C. parvum are the most common, causing > 90% of human cryptosporidiosis cases [5]. Other species including C. meleagridis, C. ubiquitum, and C. cuniculus have emerged as zoonotic pathogens and have received more attention based on their higher number of sporadic cases and resultant human cryptosporidiosis outbreaks [5]. Contact with animals is a risk factor for disease [7].
Most current genotyping studies are based on the sequence analysis of 243 bp of the ITS region, regarded as the standard method for the detection and identification of E. bieneusi genotypes [8]. To date, more than 400 ITS genotypes have been reported that can unambiguously be placed into 11 distinct clades by phylogenetic analysis [9, 10]. A large group (Group 1) comprises genotypes from both humans and animals, showing its zoonotic nature; the other eight groups (Groups 2 to 11) mainly include genotypes from specific hosts or wastewater, showing host adaption [10, 11]. To improve our understanding of the epidemiology of human microsporidiosis and strengthen the knowledge of E. bieneusi populations and their human transmission, epidemiological surveys should focus on genotyping E. bieneusi isolates from animal hosts that are in close contact with humans.
Asiatic black bears are frequently found in Chinese zoos as commercial and ornamental animals, and are therefore in close contact with zoo-keepers [12]. As a result of the high-density feeding environment in zoos and potential exposure to bear feces, infective spores of E. bieneusi from Asiatic black bears pose a potential risk to other animals and public health [13]. Currently, only two reports described E. bieneusi infections in these animals in China and no information about prevalence and genotypes of Cryptosporidium infection in Asiatic black bears in mainland of China is available [12, 13]. The health of bears is thus important in terms of management objectives and public health. The aim of this study was to determine the prevalence of natural Cryptosporidium and E. bieneusi infections in bears from the Heilongjiang and Fujian Province, China, to genotype Cryptosporidium and E. bieneusi isolates by sequence analysis of PCR products of the SSU rRNA and the ITS region of the rRNA gene, respectively. We further assessed potential zoonotic transmission by phylogenetic analysis based on the neighboring-joining method.
Results
Occurrence of Cryptosporidium and E. bieneusi
A total of 4/218 (1.8, 95% CI: 0–3.6%) and 14/218 (6.4, 95% CI: 3.1–9.7%) fecal specimens were positive for Cryptosporidium and E. bieneusi, respectively, and the differences between the overall prevalence values of the two pathogens were significant (P = 0.03, χ2 = 5.80). The prevalences of Cryptosporidium in bears was 2.8% (1/36) in Heilongjiang and 1.6% (3/182) in Fujian, while the prevalence of E. bieneusi in bears was 6.4% (14/218) with 22.2% (8/36) in Heilongjiang and 3.3% (6/182) in Fujian (Table 1). There were significant differences in the infection rates of E. bieneusi amongst bears from Heilongjiang and Fujian (P = 0.006), but no differences in Cryptosporidium infection in the two farms was observed (P > 0.05).
Genotyping and subtyping of Cryptosporidium
All four Cryptosporidium-positive specimens were successfully sequenced at the SSU rRNA locus. By sequence analysis, C. andersoni (n = 3) and Cryptosporidium rat genotype IV (n = 1) were identified. All three sequences of C. andersoni had 100% homology and were identical to DQ989573 from bactrian camels in Henan, China. The Cryptosporidium rat genotype IV sequence (MN726617) obtained was not reported previously and had 99.3% homology to the sequence (MG917670) of the isolate from brown rats in Heilongjiang, China.
All three C. andersoni-positive isolates were further subtyped through the amplification of four minisatellite/microsatellite loci (MS1, MS2, MS3 and MS16) and all were successfully amplified and sequenced. Only one MLST subtype (A4, A4, A4, A1) termed was identified. All four microsatellite/minisatellite sequences identified were described previously.
In this study, C. andersoni was only found in male animals aged ≥5 years, whilst Cryptosporidium rat genotype IV was identified in young male bears from Fujian (Table 1).
Genotyping of E. bieneusi
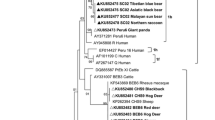
By sequence analysis of the ITS region of 14 E. bieneusi isolates obtained in this study, two known genotypes were identified including D (n = 4) and SC02 (n = 10). The distributions of E. bieneusi genotypes in animals was dependent on age, gender, and feeding mode (Table 1). Genotype D was found solely in Fujian and SC02 were found both in Heilongjiang and Fujian. Genotypes were classified in Group 1, which represents a zoonotic group (data not show).
Discussion
Cryptosporidium has only been reported in two captive Malayan sun bears (Helarctos malayanus) from Taiwan, China, in a black bear from Virginia, USA [14,15,16] and in 35 brown bears from the Slovak Republic [17]. The Cryptosporidium prevalence of 2.4% in this study was lower than the 55.6% reported in the Slovak Republic [17]. To date, only three studies of E. bieneusi in bears have been reported, two of which are from China [12, 13], with one study from the USA [18]. In China, the prevalence was 19.75% (80/405) and 27.4% (29/106) in Yunnan and Southwestern regions, respectively [12, 13], both of them were significantly lower than the prevalence of 40% (2/5) observed in wild black bears in New York City [11]. In fact, the bears in the studies from China are captive, while the animals in the study from USA are wild bears. Meanwhile, the number of the wild bears are limited (only five wild bears were examined). Here, the prevalence of E. bieneusi in bears was 6.4% (14/218) with 22.2% (8/36) in Heilongjiang and 3.3% (6/182) in Fujian. The observed differences in prevalence of E. bieneusi among different bears may be explained by variations in feeding density, geography, management systems, sample sizes, and climate, however, the possible influence of the mentioned factors above on the prevalence remains entirely unexplored, and further studies are still necessary to elucidate this aspect.
To date, only two studies have characterized Cryptosporidium in bears and only C. parvum and Cryptosporidium bear genotype have been identified in those animals [15, 16]. Xiao et al. (2000) identified the Cryptosporidium bear genotype in black bears and showed its close relationship to C. canis at the SSU rRNA and HSP-70 loci [15]. Duncan et al. (1999) detected C. parvum in tissue sections from the small intestine of black bear cubs found dead in Virginia (USA) [16]. In the present study, C. andersion and Cryptosporidium rat genotype IV were identified in bears, and C. andersion had the highest frequency and widest distribution. To the best of our knowledge, this was the first identification of C. andersion and Cryptosporidium rat genotype IV in bears, which provides useful information on the molecular epidemiology and control of Cryptosporidium infections in animals.
Although C. andersoni is widely considered a cattle-specific Cryptosporidium species, it’s occurrence in other animals and humans is increasing [18, 19]. To date, C. andersoni has been detected in cattle, yaks, sheep, goats, deer, rodents, houses, non-human primates, camels, and giant pandas [18, 20,21,22,23,24,25,26,27,28,29]. Contrasting with its occasional detection in other animals, C. andersoni is the common Cryptosporidium species in cattle, which appear its natural host [18]. In fact, C. andersoni is commonly observed in adult cattle and has been associated with gastritis, reduced milk yield, and poor weight gain. A limited number of studies have reported C. andersoni infections in humans from France, Malawi, Iran, England and Australia [19, 30,31,32,33,34]. However, recent studies reported that C. andersoni was found in 78 diarrheal patients from southern Assam, India [35], and 34 and 21 patients with diarrhea in Shanghai and Jiangsu of China [19, 36].
With the development of Cryptosporidium subtyping tools, more and more C. andersoni isolates were subtyped with a multilocus sequence typing (MLST) tool based on the MS1, MS2, MS3, and MS16 loci in China, those isolates were mainly from cattle as well as from yaks, sheep, horses, golden takin, camels and non-human primates [18, 24,25,26,27,28,29]. Sixteen MLST subtypes were identified. Amongst them A4, A4, A4, A1 was the predominant subtype in cattle from Henan, Jilin, Guangxi, Heilongjiang, Sichuan, and Shaanxi of China, and was found in non-human primates and horses from Qinling Mountains and Heilongjiang, China, respectively [18, 24, 27]. In this study, all three isolates of C. andersoni identified in bears had the A4, A4, A4, A1 subtype. The results showed that C. andersoni isolates in bears were likely to be transmitted from other animals, particularly cattle, but the real source of infection and transmission require further analysis.
Cryptosporidium rat genotype IV, previously named the Cryptosporidium W19 or Cryptosporidium W19 variant, have been recorded in rats including Asian house rats, brown rats, Edward’s long-tailed rats and muridae [37]. Meanwhile, epidemiology data suggests contamination of the water supplies evidenced by their detection in streams in the USA and raw water in the UK and China [38,39,40]. No occurrences of Cryptosporidium rat genotype IV in any animal species other than rats have been reported. This is the first report of Cryptosporidium rat genotype IV in bears, indicating that this genotype has a broader range of reservoir hosts than initially anticipated. The potential of Cryptosporidium rat genotype IV to cause disease in humans or livestock is unknown, and the source of Cryptosporidium rat genotype IV infection and its transmission dynamics now require further investigation. This will reveal the cross-species transmission potential of Cryptosporidium rat genotype IV in rats and other animals, including bears in China.
To date, 20 genotypes (D, SC01, SC02, CHB1, horse2, ABBA, ABB2 and MJ1 to MJ13) of E. bieneusi have been identified in bears, of which only genotype D has been shown to have human infective potential [11,12,13]. In this study, two known E. bieneusi genotypes, D and SC02, were identified. Genotype SC02 was identified in 71.4% of E. bieneusi isolates and was found in both Heilongjiang and Fujian. This genotype was first identified in captive wildlife (Malayan sun bear, Tibetian blue bear and Asiatic black bear) from Sichuan, China in 2016 [41]. A variety of wild animals, including Red-Bellied Tree Squirrels, horses, captive Eurasian wild boars, non-human primates, and captive giant pandas were later found to be infected [42,43,44,45]. Several studies suggest that bears are a major host for genotype SC02 [12, 13]. System development analysis showed that the genotype has the ability to infect humans, although no reported cases have been documented. Genotype D was found in four bears from Fujian, and this genotype is responsible for most human infections and identified in ≥40 countries [4]. It has been isolated from at least 15 species of animals and some water bodies, and is considered to harbor the highest potential for zoonotic transmission [9, 46]. The occurrence of E. bieneusi genotype D in bears suggests its significant zoonotic potential and threat to human health.
Conclusions
This study demonstrated the occurrence of Cryptosporidium and E. bieneusi in bears in the Heilongjiang and Fujian Provinces of China and provides molecular characterizations of C. andersoni and Cryptosporidium rat genotype IV in bears. Genotypes D and SC02 were identified in bears, suggesting their zoonotic potential and threat to human health.
Methods
Collection of fecal specimens
From May 2015 to December 2017, a total of 218 fresh fecal specimens (approximately 50 g) were collected from bears in the Heilongjiang (zoo) and Fujian (farm) Provinces of China (Table 1). All fecal specimens were collected from the ground immediately after defecation using a sterile disposable latex glove and each was placed in a labeled sterile bag. To avoid duplicate sampling, each individual animal was identified according to body characteristics such as color and size. The number of collected specimens accounted for approximately 30% of adult or young bears in the farm. All of the specimens were transported to the laboratory in a cooler with ice packs (< 48 h) and stored at 4 °C until processing (< 12 h). The ages of the animals ranged from 1 to 23 years. All animals had no apparent clinical symptoms at the time of sampling.
DNA extraction
Ten grams of feces of each sample were thoroughly mixed with 30 mL distilled water and filtrates were concentrated by centrifugation at 1500 g for 10 min. Genomic DNA was directly extracted from 200 mg of each processed fecal specimen using a QIAamp DNA stool mini kit (QIAgen, Hilden, Germany), according to the manufacturer-recommended procedures (to obtain high yields of DNA, the lysis temperature was increased to 95 °C). Extracted DNA (200 μL) from each sample was transferred to Eppendorf tubes and stored at − 20 °C prior to PCR amplification.
Genotyping and subtyping of Cryptosporidium and E. bieneusi
Cryptosporidium in the fecal specimens were identified by nested PCR amplification of the SSU rRNA gene fragment of ~ 830 bp and primers and cycle parameters were designed by Xiao et al. (1999) [47]. Meanwhile, all C. andersoni-positive isolates were further subtyped through the amplification of four minisatellite/microsatellite markers by nested PCR, respectively, including MS1 coding for hypothetical protein (550 bp), MS2 coding for 90 kDa heat shock protein (450 bp), MS3 coding for hypothetical protein (530 bp), and MS16 coding for leucine rich repeat family protein (590 bp). PCR primers and amplification conditions were performed as previously described [48]. Likewise, all DNA preparations were analyzed for the presence of E. bieneusi by amplifying a ~ 390-bp region of the rRNA gene. The PCR primers and amplification conditions were performed as previously described [49]. TaKaRa Taq DNA Polymerase (TaKaRa Bio Inc., Tokyo, Japan) was used for all PCR amplifications. Positive chicken-derived C. bailey DNA for Cryptosporidium and deer-derived BEB6 DNA for E. bieneusi) and negative controls (2 μL distilled water) were included in all PCRs. Secondary PCR products were subjected to 1.5% agarose gel electrophoresis and visualized by staining with GelRed (Biotium Inc., Hayward, CA).
DNA sequencing and analysis
All nested PCR products were sequenced using the same PCR primers used for secondary PCRs on an ABI PRISM™ 3730 DNA Analyzer (Applied Biosystems, Carlsbad, CA, USA), using a BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems). The accuracy of the sequencing data was confirmed by PCR sequencing in both directions. The species or genotypes of Cryptosporidium and the genotypes of E. bieneusi were identified by comparison to the nucleotide sequences obtained with published GenBank sequences using the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi. nlm.nih.gov/Blast.cgi) and ClustalX 1.83 (http:// www.clustal.org/), respectively.
Statistical analysis
Data entry and analysis were performed using Social Sciences (SPSS) 19.0 software. The significance of differences in infection proportions was evaluated using a Pearson’s Chi-square test, whilst the Fisher Exact test was used when more than 20% of cells in the contingency tables had expected frequencies ≤5. P-values ≤0.05 were deemed significant. The 95% confidence intervals (95% CI) for prevalence were calculated based on the Poisson distribution.
Availability of data and materials
All data generated and/or analyzed during this study are included in this published manuscript. The novel Cryptosporidium rat genotype IV nucleotide sequence obtained has been deposited in GenBank database under the following accession numbers: MN726617.
Abbreviations
- CI:
-
Confidence intervals
- ITS:
-
Internal transcribed spacer
- MLST:
-
Multilocus sequence typing
- PCR:
-
Polymerase chain reaction
- SSU rRNA:
-
Small subunit of nuclear ribosomal RNA
References
Ryan U, Fayer R, Xiao L. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology. 2014;141(13):667–85.
Fayer R, Santin-Duran M. Epidemiology of microsporidia in human infections. In: Weiss LM, Becnel JJ, editors. Microsporidia: pathogens of opportunity. 1st ed. Chichester: Wiley; 2014. https://doi.org/10.1002/9781118395264.ch3.
Checkley W, White AC, Jr Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect Dis. 2015;15(1):85–94.
Matos O, Lobo ML, Xiao L. Epidemiology of Enterocytozoon bieneusi infection in humans. J Parasitol Res. 2012;2012:981424.
Feng Y, Ryan UM, Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018;34(11):997–1011.
Holubová N, Zikmundová V, Limpouchová Z, Sak B, Konečný R, Hlásková L, et al. Cryptosporidium proventriculi sp. n. (Apicomplexa: Cryptosporidiidae) in Psittaciformes birds. Eur J Protistol. 2019;69:70–87.
Conrad CC, Stanford K, Narvaez-Bravo C, Callaway T, McAllister T. Farm fairs and petting zoos: a review of animal contact as a source of zoonotic enteric disease. Foodborne Pathog Dis. 2017;14(2):59–73.
Santín M, Fayer R. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J Eukaryot Microbiol. 2009;56(1):34–8.
Zhao W, Wang J, Ren G, Yang Z, Yang F, Zhang W, et al. Molecular characterizations of Cryptosporidium spp. and Enterocytozoon bieneusi in brown rats (Rattus norvegicus) from Heilongjiang Province, China. Parasit Vectors. 2018;11(1):313.
Zhang Y, Koehler AV, Wang T, Haydon SR, Gasser RB. New operational taxonomic units of Enterocytozoon in three marsupial species. Parasit Vectors. 2018;11(1):371.
Guo Y, Alderisio KA, Yang W, Cama V, Feng Y, Xiao L. Host specificity and source of Enterocytozoon bieneusi genotypes in a drinking source watershed. Appl Environ Microbiol. 2014;80(1):218–25.
Wu J, Han JQ, Shi LQ, Zou Y, Li Z, Yang JF, Huang CQ, Zou FC. Prevalence, genotypes, and risk factors of Enterocytozoon bieneusi in Asiatic black bear (Ursus thibetanus) in Yunnan Province, Southwestern China. Parasitol Res. 2018;117(4):1139–45.
Deng L, Li W, Zhong Z, Gong C, Cao X, Song Y, et al. Multi-locus genotypes of Enterocytozoon bieneusi in captive Asiatic black bears in southwestern China: high genetic diversity, broad host range, and zoonotic potential. PLoS One. 2017;12:e0171772.
Kváč M, McEvoy J, Stenger B, Clark M. Cryptosporidiosis in other vertebrates. In: Cacciò SM, Widmer G, editors. Cryptosporidium: parasite and disease. Vienna: Springer; 2014. p. 237–323.
Xiao L, Limor JR, Sulaiman IM, Duncan RB, Lal AA. Molecular characterization of a Cryptosporidium isolate from a black bear. J Parasitol. 2000;86(5):1166–70.
Duncan RB, Caudell D, Lindsay DS, Moll HD. Cryptosporidiosis in a black bear in Virginia. J Wildl Dis. 1999;35(2):381–3.
Ravaszova P, Halanova M, Goldova M, Valencakova A, Malcekova B, Hurníková Z, et al. Occurrence of Cryptosporidium spp. in red foxes and brown bear in the Slovak Republic. Parasitol Res. 2012;110(1):469–71.
Wang R, Zhao G, Gong Y, Zhang L. Advances and perspectives on the epidemiology of bovine Cryptosporidium in China in the past 30 years. Front Microbiol. 2017;8:1823.
Jiang Y, Ren J, Yuan Z, Liu A, Zhao H, Liu H, et al. Cryptosporidium andersoni as a novel predominant Cryptosporidium species in outpatients with diarrhea in Jiangsu Province, China. BMC Infect Dis. 2014;14:555.
Huang J, Zhang Z, Zhang Y, Yang Y, Zhao J, Wang R, et al. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in deer in Henan and Jilin, China. Parasit Vectors. 2018;11(1):239.
Osman M, El Safadi D, Benamrouz-Vanneste S, Cian A, Moriniere R, Gantois N, et al. Prevalence, transmission, and host specificity of Cryptosporidium spp. in various animal groups from two French zoos. Parasitol Res. 2017;116(12):3419–22.
Deng L, Li W, Zhong Z, Gong C, Cao X, Song Y, et al. Occurrence and genetic characteristics of Cryptosporidium hominis and Cryptosporidium andersoni in horses from southwestern China. J Eukaryot Microbiol. 2017;64(5):716–20.
Hijjawi N, Mukbel R, Yang R, Ryan U. Genetic characterization of Cryptosporidium in animal and human isolates from Jordan. Vet Parasitol. 2016;228:116–20.
Liu A, Zhang J, Zhao J, Zhao W, Wang R, Zhang L. The first report of Cryptosporidium andersoni in horses with diarrhea and multilocus subtype analysis. Parasit Vectors. 2015;8:483.
Zhao GH, Du SZ, Wang HB, Hu XF, Deng MJ, Yu SK, et al. First report of zoonotic Cryptosporidium spp., Giardia intestinalis and Enterocytozoon bieneusi in golden takins (Budorcas taxicolor bedfordi). Infect Genet Evol. 2015;34:394–401.
Wang R, Li G, Cui B, Huang J, Cui Z, Zhang S, et al. Prevalence, molecular characterization and zoonotic potential of Cryptosporidium spp. in goats in Henan and Chongqing, China. Exp Parasitol. 2014;142:11–6.
Du SZ, Zhao GH, Shao JF, Fang YQ, Tian GR, Zhang LX, et al. Cryptosporidium spp., Giardia intestinalis, and Enterocytozoon bieneusi in captive non-human Primates in Qinling Mountains. Korean J Parasitol. 2015;53(4):395–402.
Liu X, Zhou X, Zhong Z, Deng J, Chen W, Cao S, et al. Multilocus genotype and subtype analysis of Cryptosporidium andersoni derived from a Bactrian camel (Camelus bactrianus) in China. Parasitol Res. 2014;113(6):2129–36.
Wu Y, Chang Y, Zhang X, Chen Y, Li D, Wang L, et al. Molecular characterization and distribution of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi from yaks in Tibet, China. BMC Vet Res. 2019;15(1):417.
Guyot K, Follet-Dumoulin A, Lelievre E, Sarfati C, Rabodonirina M, Nevez G, et al. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J Clin Microbiol. 2001;39(10):3472–80.
Leoni F, Amar C, Nichols G, Pedraza-Diaz S, McLauchlin J. Genetic analysis of Cryptosporidium from 2414 humans with diarrhoea in England between 1985 and 2000. J Med Microbiol. 2006;55(Pt 6):703–7.
Morse TD, Nichols RA, Grimason AM, Campbell BM, Tembo KC, Smith HV. Incidence of cryptosporidiosis species in paediatric patients in Malawi. Epidemiol Infect. 2007;135(8):1307–15.
Agholi M, Hatam GR, Motazedian MH. HIV/AIDS-associated opportunistic protozoal diarrhea. AIDS Res Hum Retrovir. 2013;29(1):35–41.
Waldron LS, Dimeski B, Beggs PJ, Ferrari BC, Power ML. Molecular epidemiology, spatiotemporal analysis, and ecology of sporadic human cryptosporidiosis in Australia. Appl Environ Microbiol. 2011;77(21):7757–65.
Hussain G, Roychoudhury S, Singha B, Paul J. Incidence of Cryptosporidium andersoni in diarrheal patients from southern Assam, India: a molecular approach. Eur J Clin Microbiol Infect Dis. 2017;36(6):1023–32.
Liu H, Shen Y, Yin J, Yuan Z, Jiang Y, Xu Y, et al. Prevalence and genetic characterization of Cryptosporidium, Enterocytozoon Giardia and Cyclospora in diarrheal outpatients in China. BMC Infect Dis. 2014;14:25.
Zhao W, Zhou H, Huang Y, Xu L, Rao L, Wang S, et al. Cryptosporidium spp. in wild rats (Rattus spp.) from the Hainan Province, China: molecular detection, species/genotype identification and implications for public health. Int J Parasitol Parasites Wildl. 2019;9:317–21.
Jiang J, Alderisio KA, Xiao L. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl Environ Microbiol. 2005;71(8):4446–54.
Chalmers RM, Robinson G, Elwin K, Hadfield SJ, Thomas E, Watkins J, et al. Detection of Cryptosporidium species and sources of contamination with Cryptosporidium hominis during a waterborne outbreak in north West Wales. J Water Health. 2010;8(2):311–25.
Feng Y, Li N, Duan L, Xiao L. Cryptosporidium genotype and subtype distribution in raw wastewater in Shanghai, China: evidence for possible unique Cryptosporidium hominis transmission. J Clin Microbiol. 2009;47(1):153–7.
Li W, Deng L, Yu X, Zhong Z, Wang Q, Liu X, et al. Multilocus genotypes and broad host-range of Enterocytozoon bieneusi in captive wildlife at zoological gardens in China. Parasit Vectors. 2016;9(1):395.
Li W, Zhong Z, Song Y, Gong C, Deng L, Cao Y, et al. Human-pathogenic Enterocytozoon bieneusi in captive Giant pandas (Ailuropoda melanoleuca) in China. Sci Rep. 2018;8(1):6590.
Li W, Deng L, Wu K, Huang X, Song Y, Su H, et al. Presence of zoonotic Cryptosporidium scrofarum, Giardia duodenalis assemblage a and Enterocytozoon bieneusi genotypes in captive Eurasian wild boars (Sus scrofa) in China: potential for zoonotic transmission. Parasit Vectors. 2017;10(1):10.
Deng L, Li W, Zhong Z, Gong C, Liu X, Huang X, et al. Molecular characterization and multilocus genotypes of Enterocytozoon bieneusi among horses in southwestern China. Parasit Vectors. 2016;9(1):561.
Deng L, Li W, Yu X, Gong C, Liu X, Zhong Z, et al. First report of the human-pathogenic Enterocytozoon bieneusi from red-bellied tree squirrels (Callosciurus erythraeus) in Sichuan, China. PLoS One. 2016;11(9):e0163605.
Wang H, Liu Q, Jiang X, Zhang Y, Zhao A, Cui Z, et al. Dominance of zoonotic genotype D of Enterocytozoon bieneusi in bamboo rats (Rhizomys sinensis). Infect Genet Evol. 2019;73:113–8.
Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ, et al. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. 1999;65(4):1578–83.
Feng Y, Yang W, Ryan U, Zhang L, Kvác M, Koudela B, et al. Development of a multilocus sequence tool for typing Cryptosporidium muris and Cryptosporidium andersoni. J Clin Microbiol. 2011;49(1):34–41.
Buckholt MA, Lee JH, Tzipori S. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl Environ Microbiol. 2002;68(5):2595–9.
Acknowledgements
Not applicable.
Funding
This work was supported partially by the Fundamental Research Funds for the West Light Talents Training Programme of 2018 and the Open Subject of Key Laboratory of Parasite and Vector Biology, MOPH (WSBKFKT-201802). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author information
Authors and Affiliations
Contributions
W-S L and WZ contributed to the conception and design of the experiments. S-N W, YS, W-S L and GL were involved in the sample collection. S-N W, YS and H-H Z performed the experiments. WZ, W-S L and MQ contributed to analysis of results and data interpretation. WZ and W-S L contributed to manuscript drafting. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was performed in accordance with the Law of the People’s Republic of China on the Protection of Wildlife of 1989. The research protocol was reviewed and approved by the Research Ethics Committee of Northeast Forestry University. Prior to the collection of fecal specimens, permission was obtained from zoo-keeper and bear farm managers for animal involvement. No animals were injured during the study protocols.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wang, SN., Sun, Y., Zhou, HH. et al. Prevalence and genotypic identification of Cryptosporidium spp. and Enterocytozoon bieneusi in captive Asiatic black bears (Ursus thibetanus) in Heilongjiang and Fujian provinces of China. BMC Vet Res 16, 84 (2020). https://doi.org/10.1186/s12917-020-02292-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-020-02292-9