Abstract
Background
The GABARAP family members (GABARAP, GABARAPL1/GEC1 and GABARAPL2 /GATE-16) are involved in the intracellular transport of receptors and the autophagy pathway. We previously reported that GABARAPL1 expression was frequently downregulated in cancer cells while a high GABARAPL1 expression is a good prognosis marker for patients with lymph node-positive breast cancer.
Methods
In this study, we asked using qRT-PCR, western blotting and epigenetic quantification whether the expression of the GABARAP family was regulated in breast cancer by epigenetic modifications.
Results
Our data demonstrated that a specific decrease of GABARAPL1 expression in breast cancers was associated with both DNA methylation and histone deacetylation and that CREB-1 recruitment on GABARAPL1 promoter was required for GABARAPL1 expression.
Conclusions
Our work strongly suggests that epigenetic inhibitors and CREB-1 modulators may be used in the future to regulate autophagy in breast cancer cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Autophagy is a cell process which regulates cell homeostasis and survival by inducing the degradation and recycling of intracellular components like protein aggregates or organelles (such as damaged mitochondria) [1]. This mechanism involves more than 40 proteins required for : i) the formation of autophagosomes, ii) the fusion of autophagosomes with lysosomes and/or, iii) the regulation of autophagy flux. The role of autophagy in tumorigenesis is controversial, even though evidence indicated that autophagy is often downregulated in cancer cells. On one hand, autophagy might protect against transformation of non tumoral cells into cancer cells and in the other hand, a loss of autophagy might help the cancer cells to escape from type II cell death (also called autophagy cell death) [2, 3]. Since induction of autophagy in cancer cells has also been described to confer resistance to chemotherapeutive agents, the co-administration of both alkyling agents and autophagy inhibitors (such as hydroxychloroquine) could improve the anti-tumoral response in these resistant cells [4]. Among the proteins involved in autophagy, two subfamilies of homologs of the yeast Atg8 (Autophagy-related 8) have been described in mammals: i) the MAP-LC3s (Microtubule-associated protein Light Chain 3) including LC3A, LC3B and LC3C and, ii) the GABARAP (GABAA-receptor-associated protein) family. The latter comprises 3 members: GABARAP, GABARAPL1/GEC1/ATG8L (GABARAP-like protein 1/guinea-pig endometrial glandular epithelial cells-1/Atg8-like protein) and GABARAPL2/GATE-16 (GABARAP-like protein 2/Golgi-associated ATPase enhancer of 16 kDa).
GABARAP, GABARAPL1 and GABARAPL2 genes are located on the human chromosomes, 17p13.12, 12p12.3 and 16q22.3 respectively, and are differentially expressed in normal and pathological tissues. The GABARAP gene has been described to be highly expressed in endocrine tissues while the GABARAPL1 gene is predominantly expressed in the central nervous system but they both are underexpressed in a large variety of cancer cell lines [5]. Nevertheless, the analysis of GABARAPL1 expression in a cohort of 256 breast adenocarcinoma revealed that a low GABARAPL1 expression was correlated with a high risk of metastasis, in particular for lymph node-positive patients [6]. Despite these recent studies, the regulation of the GABARAP family is still poorly understood and the origin of their decreased expression in tumor models remains unknown.
It is now recognized that epigenetic modifications control the expression of numerous genes via the regulation of promoter accessibility to transcriptional factors. Both DNA methylation and histone modifications affect the level of chromatin compaction and it has been described that epigenetically-mediated aberrant silencing of genes are an important factor in the pathogenesis of cancers including breast cancers (BC) [7, 8] Indeed, epigenetic modifications can regulate the expression of a large panel of genes involved in the hallmarks of cancer, such as apoptosis, cell signaling, invasion and proliferation. For example, the detection of the promoter methylation of the tumor suppressor gene BRCA1, which is frequent in BC and is associated with a decrease of BRCA1 expression, can help to predict the response to conventional chemotherapies in triple negative BC patients [9]. DNA methylation consists on the addition of a methyl group on a cytosine in CpG islands. It is catalyzed by DNA methyl transferases (DNMTs) and is unfavorable to transcription. Following DNA replication and formation of hemi-methylated DNA, the conservation of DNA methylation on the neo-synthesized strand, is mainly processed by DNMT1 using the parental strand as a model. This DNA methylation conservation is called maintaining or inheritance DNA methylation. On the opposite, de novo DNA methylation referred to the addition of DNA methylation on both strands of DNA on previously unmethylated loci is catalyzed by both DNMT3A and DNMT3B.
Besides DNA methylation, post-translational modifications of histones are also frequently associated to the regulation of gene expression in cancers. Histones are associated as octamers in nucleosomes (dimer of H2A, H2B, H3 and H4 and a loop of 126 pb DNA) whose compaction is regulated by post-translational modifications such as phosphorylation, methylation or acetylation. The local sum of these modifications is called the histone code and determine the status of local chromatin compaction (for a review see [10]). Histone methyl transferases (HMTs) or histone demethylases (HDMs) respectively catalyze the methylation or demethylation of histones leading to different effects on transcription. For example, H3K9me or K3K27me are negative marks while H3K4me is favorable to transcription. Acetylation, the most studied histone modification, is processed by histone acetyl transferases (HATs) and is associated with a local relaxed chromatin and is therefore favorable to gene expression. On the opposite, the removal of acetyl groups from histones, which is catalyzed by histone deacetylases (HDACs), contributes to gene silencing.
Some recent studies also revealed that epigenetic modifications can regulate autophagy gene expression as well as autophagy levels in both normal and cancer cells. For example, HDACs play an essential role in the regulation of autophagy: HDAC1 inhibition favors the conversion of the soluble LC3B form (LC3B-I) to the membrane-bound form of LC3B (LC3B-II), while the presence of H4K16ac (catalyzed by hMOF (human ortholog of drosophila males absent in the first)) in some ATGs genes, is associated to a decrease of expression of these genes [11, 12]. Moreover, HDAC6, an HDAC mainly localized in the cytosol, has also been described to be involved in the transport and maturation of autophagosomes [13]. DNA methylation is also involved in autophagy regulation as hypermethylation of several ATG genes has been described in various cancers [13, 14]. For example, methylation of BECN-1, a tumor suppressor gene, has been observed in BC, while methylation of ATG16L2, LC3A, ULK2, or BNIP3 has been suggested to be involved in the down-regulation of autophagy in other cancers [15–19].
In order to characterize the regulation of GABARAP gene family expression in cancer cells, we analyzed their expression and the epigenetic modifications in the promoters of these genes in in vitro human BC cell models. Our data demonstrated that GABARAPL1 expression is decreased in BC patients and BC cell line models. Moreover, both DNA methylation and deacetylation of histone H3 in GABARAPL1 promoter were observed in these BC cell line models while the inhibition of DNMTs and HDACs using specific inhibitors restored GABARAPL1 expression in these cells. These data suggest that DNMTi (DNMT inhibitors) and HDACi (HDAC inhibitors) could be used in the future to modulate autophagy levels in BC cells.
Methods
Ethic statement
Human samples were collected according to French laws and the recommendations of the French National Committee of Ethics. Indeed, this study has been approved by of the scientific committee of “the tumorothèque régionale de Franche-Comté BB-0033-00024”. The samples and the medical history of patients were encoded to protect patients confidentiality and used under protocols approved by the recommendations of the French national Committee of Ethics. All human samples were collected by Pr. Severine Valmary-Degano (Centre Hospitalo-Universitaire, Besançon, France) at the “Tumorothèque régionale de Franche-Comté BB-0033-00024”. Collection of samples and their use (AC-2010-1163) for studies (approved by the scientific committee of “the tumorothèque régionale de Franche-Comté BB-0033-00024”) have been approved by the French “ministère de la recherché” and by the CPP EST II.We obtained all necessary consents from any patients involved in the study.
Quantitative RT-PCR
RNA was isolated from cells and frozen tissues using Tri Reagent (Molecular Research Center, TR-118) as described by the manufacturer. Reverse transcription were performed using M-MLV (Sigma-Aldrich, M-1302) reverse transcriptase and 1.5 μg total RNA according to manufacturer’s instructions (Sigma-Aldrich). Quantitative PCR (qPCR) were done in duplicate using the Step one Real-Time PCR system (Applied Biosystems), Power SYBR Green PCR Master Mix (Applied Biosystems, 4367659), according to manufacturer’s instructions and primers specific of GABARAP (F: 5′-GCCTTTCCCATCCTGCTGTA-3′ and R: 5′-AGGAGGGGATTGCTGGGTTCT-3′) ; GABARAPL1 (F: 5′-CCCTCCCTTGGTTATCATCCA-3′ and R: 5′-ACTCCCACCCCACAAAATCC-3′) and GABARAPL2 (F: 5′-AAATATCCCGACAGGGTTCC-3′ and R: 5′-CAGGAAGATCGCCTTTTCAG-3′). H3B2 was used as an housekeeping gene (F: 5′-GCTAGCTGGATGTCTTTTGG-3′ and R: 5′-GTGGTAAAGCACCCAGGAA-3′) as previously described [20].
Cell culture
MCF-7 and MDA-MB-453 cell lines were obtained from ATCC (HTB-130 and HTB-22) and grown in DMEM 1 g/L glucose (Dominique Dutscher, L0066) containing fetal calf serum (5 %) (Dominique Dutscher, S1810), penicillin (50 U/ml) (Dominique Dutscher, L0018), streptomycin (50 μg/ml), and amphotericin B (1.25 μg/ml) (PAA, P11-001) at 37 °C in 5 % CO2, and routinely used at 70–80 % confluence. When indicated, cells were exposed to 2 μM 5-aza-CdR (A3656, Sigma-Aldrich) for 48 h and 400 nM TSA (T8552, Sigma-Aldrich) for 16 h.
Luciferase activity
GABARAPL1 promoter fragments were obtained by PCR using the following primers–336 F: 5′-GCTGGATCCCAACCAGCAGGA-3′, −659 F: 5′-GTCAGGCTGGTCTCGAACTC-3′ and +241 R: 5′-GGGATGCACCGCAGGGCTTCC-3′ and then cloned into the pGL3 basic plasmid. 5000 cells were seeded in 96 multiwell dishes and cells were transfected with pGL3 plasmids, co-transfected with the pCDNA3.1-CREB-1 vector (kindly provided by Vincent Coulon, Montpellier, France) or treated with 10 μM forskolin. Luciferase expression was measured using the Luciferase Assay System kit (E1500, Promega) according to the manufacturer’s recommendations.
Epigenetics
gDNA was extracted using the NucleoSpin® Tissue kit (740952, Macherey Nagel). Global DNA methylation was quantified using the MethylFlash methylated DNA quantification kit (P-1034, Epigentek, France). Methyl DNA collection were performed using the “Methyl-Collector Ultra kit” (55005, Active Motif). Histone 3 acetylation was quantified using the EpiQuik Tissue Acetyl-Histone H3 ChIP Kit (P-2012, Epigentek). ChIP was performed using the ChIP-IT High Sensitivity kit (53040, Active Motif) with ChIP grade antibodies (Table 1). All these kits were used according to the manufacturer’s instructions. Primers used in this study were designed with the primer3 software [21]: GABARAP (F: 5′-AAAGCCAACCGTCTTTGCTA-3′ and R: 5′-GCCACTTCCCTATTCACCAA-3′), GABARAPL1 (MC1 F: 5′-GTCAGGCTGGTCTCGAACTC-3′ and R: 5′-CGCTCCTGAACAGCAACATA-3′) and GABARAPL1 (MC2 F: 5′-AAGGAAACGCAGTGAGACAGA-3′ and R: 5′-AGCTGGGAGCACAAAAACAG-3′), GABARAPL2 (F: 5′AATTCCCCAGACTTCCCCTA-3′ and R: 5′-GGTGGCGAAGAAGTTGGTTA-3′).
Western-blotting
Cells were scraped, harvested and lysed in RIPA buffer (50 mM Tris–HCl, pH 8, 150 mM NaCl, 1 % Triton ×100, 0.5 % DOCA, 0.1 % SDS) supplemented with protease inhibitors (104 mM AEBSF, 1.5 mM pepstatin A, 1.4 mM E-64, 4 mM bestatin, 2 mM leupeptin, 80 μM aprotinin) for 30 min on ice, sonicated for 15 sec and centrifuged at 10 000 g for 10 min at 4 °C. Supernatant was used for protein quantification using the Bradford method [22] and then proteins (40 μg) were separated using SDS-PAGE gels and transferred to PVDF membranes (Bio-Rad, 162–0177) for 2 h in Tris-Glycine buffer as previously described [23]. Membranes were saturated in 0.1 % TBS-Tween 20 and 5 % nonfat milk for 1 h and then incubated with primary antibodies (Table 1) overnight at 4 °C. Membranes were washed 3 times with TBS-Tween 20 0.1 %, incubated with secondary anti-rabbit HRP conjugate or anti-mouse HRP conjugate antibody according to manufacturer’s instructions (P.A.R.I.S., BI2407 and BI2413C). The membrane was washed 3 times with TBS-0.1 % Tween 20 incubated with ECL revelation buffer (Pierce) and cheluminescence was monitored using a ChemiDocTMXRS+ (Biorad).
Immunofluorescence
CREB-1 IF was performed as precognized by the manufacturer. Briefly, cells were seeded on coverslips in 24 multiwell-plates, fixed and permeabilized for 20 min with cold methanol at–20 °C, washed 3 times with cold PBS (Phosphate buffer saline : 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4), incubated with 1 % BSA-PBS for 1 h at 37 °C and incubated overnight with the CREB-1 antibody (Table 1). The coverslips were washed 3 times for 5 min with 0.1 % Tween-PBS (T-PBS), incubated for 1 h with a goat secondary anti-mouse Alexa 555 antibody (Life technology, France) and washed 3 times for 5 min with (T-PBS). Cells were then mounted in Vectashield Hardset mounting medium (Vector Laboratories, H-1000) and analyzed using an Olympus IX81 confocal microscope (Olympus, France).
Statistics
Mean’s comparison were analyzed using a Student t-test with GraphPad Prism5 software (USA). Correlation indexes were measured using a Spearman test with ImageJ software. Significant values were highlighted in bold in each figure.
Results
GABARAP family genes are differentially expressed in human breast cancer biopsies
We first analyzed GABARAP, GABARAPL1 or GABARAPL2 mRNA expression in human BC biopsies using qRT-PCR (Fig. 1). The different BC subtypes are classified in regard of their molecular marker expression. Luminal BC, which represent 50 % of total BC and generally associated with a good prognosis, are divided in Luminal A and Luminal B BC. Both Luminal A and B BC express ERα (estrogen receptor: ER+) whereas the expression of the HER2 (HER+) (human epidermal growth factor receptor) gene was only observed in Luminal B BC. HER+ BC subtype, which represents 17 % of BC, presents an amplification of the HER2 gene without the expression of ERα (ER-). The triple negative BC (ER-/PR-/HER-) do not express ERα, PR (progesterone receptor) and HER2 and cannot be treated with specific therapies (for a review, see [24]). Our cohort comprised 5 grade I BC (ER+/PR+/HER-) and 8 grade III BC (5 ER-/PR-/HER-and 3 ER+/PR+/HER+). Our results revealed an insignificant decrease of both GABARAP and GABARAPL2 mRNA levels in grade III BC compared to non tumoral tissue (NT). More interestingly, GABARAPL1 expression was strongly decreased in BC grade III tissues (p = 0.004) versus non tumoral tissues. An inverse correlation (r = −0.57) was also observed between GABARAPL1 mRNA and tumor stage while a very poor correlation was determined between GABARAP or GABARAPL2 expression levels and the tumor grade (respectively r = −0.3 and r = −0.1).
GABARAP family expression is deregulated in breast cancers. Quantification of GABARAP, GABARAPL1, GABARAPL2 expression using qRT-PCR, in grade I + II and grade III BC biopsies compared to normal adjacent tissue. Circle: NT (non tumoral), square: grade II, triangle: grade III. Difference of expression were quantified using a t-test. The correlation between the tumor grade and gene expression was measured using a Spearman test
Study of global and local epigenetic modifications in GABARAPL1 promoter and GABARAPL1 expression in BC tissues and cell models
Since tumors are frequently associated with aberrant DNA methylation content, we quantified global DNA methylation levels using ELISA in gDNA issued from both grade III BC samples (associated with the lowest expression of GABARAPL1 mRNA, as previously described in Fig. 1 and NT tissues (Fig. 2a).7 [25, 26] As expected, several gDNA issued from grade III BC presented a lower global DNA methylation compared to gDNA issued from NT tissues. These results suggested that epigenetic modifications might occur in these tumors. To determine whether local DNA methylation was also altered in these BC samples, methylation of XIST, a X-linked gene known to be methylated in women, and GAPDH, a gene constitutively active and unmethylated, were analyzed by precipitation of methylated DNA using the methylCollector Ultra kit. As expected, methylation of XIST was observed in 100 % of both gDNA issued from NT and grade I-II BC tissues tested but lost in 3 out of 5 triple negative BC biopsies (Grade III) (Additional file 1: Figure S1). On the contrary, GAPDH was never found to be methylated in NT gDNA but was surprisingly frequently methylated in gDNA issued from BC samples (Additional file 1: Figure S1). Loss of global DNA methylation in several tissues (Fig. 2a 4 out 13 samples) and aberrant status of methylation of XIST and GAPDH (Additional file 1: Figure S1) strongly suggested that DNA methylation was deregulated in BC samples.
Detection of DNA methylation in GABARAP family promoters a Global DNA methylation quantification in BC and non tumoral biopsies using methylFlash ELISA kit. b Scheme describing the GABARAP family gene promoter structure (Methprimer) and primer localization. c Left: Descriptive of GABARAP family gene methylation using methylCollector kit ; right : examples of methylation signal observed using GABARAPL1 MC2 primers. White : absence of signal of methylation; black : signal of methylation. d Correlations (Spearman test) between expression and methylation of GABARAP, GABARAPL1 and GABARAPL2 genes. NT: non tumoral, ER +/−: status of expression of estrogen receptor α, PR+/−: status of expression of progesterone receptor, HER+/−: status of expression of Human epidermal growth factor receptor; I-III: BC grade
We next analyzed whether epigenetic regulation of the GABARAPL1 might explain its specific down-regulation in BC. GABARAP, GABARAPL1 and GABARAPL2 present CpG rich areas in their promoter (−800/+200) as predicted using the methPrimer software [27], so we analyzed the methylation status of these promoters using the methylCollector Ultra kit (Fig. 2b and c). A low methylation signal was observed in both GABARAP and GABARAPL2 promoters (MC primers) both in tumors and NT tissues while primers designed in the–800 region of GABARAPL1 (MC1) (Fig. 2c) revealed a strong signal of methylation in both NT and BC tissues. Regarding GABARAPL1, a low signal of unmethylation was also measured in NT samples but not in BC samples suggesting that hemi-methylation was lost in cancer cells (p = 0.0024) (Fig. 2c). To confirm the higher level of methylation in the GABARAPL1 promoter in BC, the same experiment was repeated using primers (MC2) designed to detect the 5′-UTR region of this gene. Results obtained with MC2 primers showed that GABARAPL1 was not or weakly methylated in NT samples but highly methylated in BC tissues (p = 0.05) (Fig. 2c). Pearson correlation analysis revealed that GABARAP or GABARAPL2 expression was not correlated with methylation status (r = −0.45, p = 0.12 and r = 0.36, p = 0.25 Fig. 2d), while GABARAPL1 expression was indeed correlated with the methylation status of the gene (r = −0.58, p = 0.03 suggesting that methylation of the GABARAPL1 promoter may explain the downregulation of GABARAPL1 expression in BC (Fig. 2d).
In order to characterize the pathway allowing epigenetic modifications to control GABARAP family expression, we first analyzed GABARAP, GABARAPL1 and GABARAPL2 mRNA levels using qRT-PCR in MCF-7 and MDA-MB-453 (BC cell lines) and MCF-10A (Breast immortalized but non tumoral cell line) cell lines (Fig. 3a). Both GABARAP (p = 0.039), GABARAPL1 (p = 0.013) and GABARAPL2 (p = 0.039) expression were decreased in MCF-7 compared to MCF-10A but while the loss of GABARAP or GABARAPL2 expression was weak (0.5-2 fold), GABARAPL1 expression was about 100-fold lower in MCF-7 compared to MCF-10A (Fig. 3a). Similar results were obtained for GABARAP, GABARAPL1 and GABARAPL2 in MDA-MB-453 cells but GABARAP did not show any significant differences in this cell line (Fig. 3a). Methylation of the GABARAP family promoters was then assessed using the MethylCollector kit in both MCF-7 and MDA-MB-453 cell lines presenting a low expression of GABARAPL1. First, a signal of methylation of GABARAP promoter was observed in MCF-7 cells but not in MDA-MB-453 cells (Fig. 3b). Regarding GABARAPL2 promoter, no methylation was detected in both cell lines. A high signal of methylation was detected in the GABARAPL1 promoter in MCF-7 and MDA-MB-453 cells using MC1 primers confirming that the region of MC1 is highly frequently methylated. As expected GABARAPL1 promoter methylation was lost in MCF-7 cells treated with 5-aza-CdRdeoxycytidine (5-aza-CdR), a DNMTi (Fig. 3b). A high signal of methylation was also detected using MC2 primers in MCF-7 cells but not in MCF-10A cells suggesting, as observed before in human BC biopsies, that GABARAPL1 methylation is predominantly observed in BC cell lines (Fig. 3c)
Epigenetic modifications in GABARAP family gene promoters. a Quantification of GABARAP, GABARAPL1, GABARAPL2 expression using qRT-PCR in BC MCF-7 and MDA-MB-453 cancer cells and MCF-10A immortalized cells. b and c GABARAP family gene methylation using methylCollector kit in MCF-7, MDA-MB-453 and MCF-10A cells. (I: input; M: methylated fraction). d Visualization of H3 deacetylation using ChIP experiment and anti-H3 acetylated (H3-ac) antibody in the GABARAP family gene in MCF-7 and MDA-MB-453 cells (I: input; IgG: negative control of IP). e Detection of DNMT1 and HDAC1 recruitment on GABARAPL1 promoter using ChIP experiment and anti-DNMT1 or anti-HDAC1 antibody (I: input; IgG : negative control)
As local DNA methylation is frequently associated to histone deacetylation, the acetylation status of histone H3 (H3-ac) was analyzed by ChIP in MCF-7 and MDA-MB-453 cells previously treated or not with trichostatin A (TSA), an inhibitor of HDACs (Fig. 3d). ChIP analysis revealed that H3 acetylation was detected in the GABARAP promoter in MCF-7 cells but not in MDA-MB-453 cells. A very low level of H3 acetylation was also observed in GABARAPL2 promoter in these both cell lines. These signals were increased following TSA treatment in MCF-7 and MDA-MB-453 cells. Similarly, no H3 acetylation signal could be detected in the GABARAPL1 promoter of BC cells but this signal was increased after TSA treatment, particularly in the MDA-MB-453 cells (Fig. 3d).
All these data (Figs. 1, 2 and 3) suggest that GABARAPL1 is the most regulated gene of the GABARAP family and that require promoter deacetylation and DNA methylation. Interestingly, both DNMT1, which predominantly catalyzes inheritance DNA methylation, and HDAC1 were detected on GABARAPL1 promoter in MCF-7 cells using ChIP experiments (Fig. 3e).
We next asked whether 5-aza-CdR or TSA could restore GABARAPL1 expression in BC cell lines. To do so, MCF-7 and MDA-MB-453 cells were treated with 5-aza-CdR or TSA and the levels of GABARAP, GABARAPL1 and GABARAPL2 mRNA were measured using qRT-PCR (Fig. 4a) while protein levels were quantified by western-blotting (Fig. 4b). All cells were also treated with MG-132, before protein extraction, to prevent the fast proteasomal degradation of GABARAPL1 which has been previously reported [28]. First we observed a not significant increase of GABARAP mRNA (p = 0.07) but a significant increase of the corresponding protein GABARAP (p = 0.007 and p = 0.02) following TSA treatment in MCF-7 and MDA-MB-453 cells. Both GABARAPL2 mRNA (p = 0.05 and p = 0.05 respectively) and GABARAPL2 protein (p = 0.045 and p = 0.0003 respectively) were increased in MCF-7 and MDA-MB-453 cells treated with TSA. No significant effect of 5-aza-CdR could be observed on GABARAP and GABARAPL2 expression (Fig. 4a). On the opposite, 5-aza-CdR treatment increased GABARAPL1 mRNA level (about 2 fold) in both cell lines (p = 0.004 and p < 0.0001 respectively) while TSA treatment increased GABARAPL1 expression of 10 to 25 fold (Fig. 4a). Moreover, GABARAPL1 expression, which was undetectable in non treated MCF-7 cells or 5-aza-CdR-treated cells, was increased following TSA treatment (Fig. 4b). A weak and diffuse band signal corresponding to the GABARAPL1 protein was also in over-exposed western-blotting using lysates of MDA-MB-453 cells treated with TSA, suggesting that GABARAPL1 might also be slightly increased in these cells after TSA treatment.
Effects of epigenetic modulators on the GABARAP family gene expression. a Effects of 5-aza-deoxycytidine (5-aza-CdR) or trichostatin A (TSA) on GABARAP family gene expression using qRT-PCR analysis. b Effects of 5-aza-CdR or TSA on GABARAP family protein expression using WB (anti-GABARAP/GABARAPL1, anti-GABARAPL2 and anti-ACTIN antibodies) in cells previously treated with MG-132. Differences were quantified using a t-test
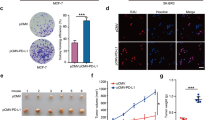
Altogether these results confirm that GABARAPL1 is the gene of the GABARAP family whose expression is the most sensitive to epigenetic regulation in BC cell lines. Since 5-aza-CdR and TSA treatments restored GABARAPL1 content, we next asked whether these compounds modulate autophagy and cell proliferation in MCF-7 cells (Additional file 2: Figure S2). Both an increase of LC3B-II (form associated to the autophagosomes) (Additional file 2: Figure S2A) and of cells with GFP-LC3 puncta (Additional file 2: Figure S2B) were observed in respectively 5-aza-CdR/ TSA treated cells and in GFP-LC3 transfected and TSA treated cells compared to control cells. Moreover, both an decrease of cell proliferation and clonogenecity were also observed in MCF-7 treated cells (Additional file 2: Figure S2C and D). These results suggest that restoration of GABARAPL1 expression might be linked to these processes although some pleiotropic effects of 5-aza-CdR and TSA treatment could also be involved [29].
GABARAPL1 expression is regulated by CREB-1
Behind epigenetic modifications of the promoter, the second step of gene regulation is the recruitment of transcriptional factors (TF). Therefore, in order to characterize the mechanisms governing GABARAPL1 expression in these cells, we cloned two fragments of the GABARAPL1 promoter (−659/+241 and −336/+241) in the pGL3 luciferase reporter plasmid. As suggested by 3 different softwares (TESS http://www.cbil.upenn.edu/tess, Cister http://zlab.bu.edu/~mfrith/cister.shtml and Patch http://www.gene-regulation.com/pub/programs.html#patch) used to predict TF binding sites, several putative CRE (cAMP Response Element) elements were identified in GABARAPL1 promoter (Fig. 5a). Indeed, following transfection of our constructions in MCF-7 cells, we observed a significant increase of luciferase signal in cells transfected with the–336/+241-GABARAPL1-promoter-pGL3 plasmid compared to the empty pGL3 vector suggesting the presence of functional regulatory elements in this region. Moreover, luciferase activity was strongly increased when cells were transfected with the–659/+241-GABARAPL1–promoter-pGL3 vector compared to the basal–336/+241-GABARAPL1-promoter-pGL3 vector suggesting that the region −659/-336 is also important for GABARAPL1 regulation (Fig. 5b). Both treatment of MCF-7 cells with forskolin, a compound known to activate CREB-1 (CRE binding protein-1), or transfection with a plasmid expressing the CREB-1 protein significantly increased luciferase activity linked to the construction–659/+241-GABARAPL1–promoter-pGL3, suggesting that CREB-1 is indeed involved in GABARAPL1 expression (Fig. 5c). The increase of luciferase activity in cells transfected with the pcDNA3.1-CREB-1 vector might be explained by the low level of endogenous CREB-1 in MCF-7 cells as observed in IF experiments (Fig. 5c) and ChIP experiment also confirmed the recruitment of CREB-1 on the–659/+241-GABARAPL1-promoter in spite of the presence of a high background noise that may be provoked by the presence of–659/+241-GABARAPL1-promoter plasmid (Fig. 5d). We next asked whether endogenous GABARAPL1 expression may be regulated by CREB-1. Treatment of MCF-7 cells with Actinomycin D revealed that the half-life of GABARAPL1 mRNA was high (about 17 h) suggesting that regulation of GABARAPL1 mRNA content is more dependent on transcription that mechanisms affecting mRNA stability (Fig. 5e) [30, 31]. Moreover, overexpression of CREB-1 in MCF-7 cells significantly increased GABARAPL1 expression but at a lower level than the ones observed in cells treated with 5-aza-CdR/TSA (Fig. 5f). Moreover, a non-significant (p = 0.066) further increase of GABARAPL1 expression was observed in cells transfected with the CREB-1 plasmid and treated with 5-aza-CdR/TSA compared to cells with 5-aza-CdR/TSA treatment alone.
GABARAPL1 expression I controlled by CREB-1. a Scheme describing the position of primers used (in regard of the putative initial transcription site (+1)) and putative CRE (CREB-1 response elements) sites in the GABARAPL1 promoter. b and c Luciferase activity measured using a Luciferase assay System Kit in MCF-7 cells transfected with empty pGL3 plasmid,−336/+241-GABARAPL1-promoter-pGL3 plasmid,–659/+241-GABARAPL1-promoter-pGL3 plasmid, pCDNA3.1-CREB-1 or treated with (10 μM) forskolin. c Bottom : expression of CREB-1 using IF in cells transfected or not with the pCDN3A.1-CREB-1 vector. d Recruitment of CREB-1 on–659/+241-GABARAPL1-promoter-pGL3 plasmid using ChIP experiment and anti-CREB-1 antibody in MCF-7 cells transfected with–659/+241-GABARAPL1-promoter-pGL3 and pCDNA3.1-CREB-1 plasmids (I: input; IgG : negative control of IP). e Half-life of GABARAPL1 mRNA using qRT-PCR following Actinomycin D treatment in MCF-7 cells. f Effects of CREB-1 overexpression (following pCDNA3.1-CREB-1 plasmid transfection) and/or 5-aza-CdR /TSA treatment on GABARAPL1 expression using qRT-PCR in MCF-7 cells. g Effects of CREB-1 overexpression (following pCDNA3.1-CREB-1 plasmid transfection) and/or 5-aza-CdR/TSA treatment on CREB-1 recruitment in GABARAPL1 promoter using ChIP experiment and an anti-CREB-1 antibody (I: input; IgG : negative control). Differences were quantified using a t-tests. GL1: GABARAPL1
These data suggest that epigenetic modifications may be an initial predominant factor allowing the recruitment of CREB-1 on GABARAPL1 promoter. These observations were partly confirmed by ChIP experiments showing an increase of CREB-1 recruitment on GABARAPL1 promoter in MCF-7 cells previously treated with 5-aza-CdR/TSA. But no significant difference could be observed between 5-aza-CdR/TSA treated cells transfected or not with the vector encoding CREB-1 (Fig. 5g).
Discussion
While the role of autophagy in tumorigenesis is still controversial, it is currently admitted that autophagy is generally reduced in cancer cells. Whereas the origin of the autophagy disruption is still unknown, mutations or loss of heterozygosity-dependent autophagy gene silencing (LC3, ATGs, UVRAG, BECN-1) have been reported in many different cancers, such as colorectal, gastric carcinoma or breast cancers [32–36]. Besides mutations, epigenetic modifications frequently occur in cancers, such as global DNA hypomethylation and/or both local hypo and hypermethylations in specific loci [37–39]. Hypermethylation of five autophagy-related genes, BECN-1, ATG16L2, ULK2, BNIP3 and a variant of LC3A, were previously reported respectively in BC, leukemia, astrocytoma, colorectal cancer and esophageal carcinoma and these data demonstrated that this hypermethylation was correlated to tumor grade [15–19]. Moreover, inhibition of the HMT EZH2 or G9a promotes autophagy in cancer cells while the use of HDACi gave contradictory results on autophagy levels in cancer cells [12, 40].
Despite a high homology between the different members of the GABARAP family (GABARAPL1 shows 87 % identity with GABARAP and 61 % with GABARAPL2), these proteins are differentially expressed during development and in adult tissues (for review see [41]) and these proteins have been described to be involved in several molecular pathways, including receptor transport and autophagy. Previous studies revealed that GABARAP and GABARAPL1 may have redundant functions in transport but experiments of invalidation/overexpression of GABARAP and GABARAPL1 showed that both are required for autophagy [42, 43]. GABARAPL1 expression has been described to be decreased in cancer cell lines. MCF-7 cells stably expressing exogenous Flag-GABARAPL1-6His presents a significantly reduced proliferation rate compared to control cells [6]. Other studies described that a high expression of this gene is associated with a positive outcome in metastatic BC [5, 6] while a low expression of GABARAPL1 was also associated with a poor outcome in kidney carcinoma patients [44]. Our results presented in this study confirmed these data by showing an inverse correlation between GABARAPL1 expression and BC grade (Fig. 1). While GABARAP and GABARAPL2 expression are also reduced in the tumor samples, this decrease was not significant. Similar differences were observed between non tumoral MCF-10A cells and MCF-7 BC cells since we showed a significant decrease of GABARAP and GABARAPL2 expression in MCF-7 cells but the highest decrease was observed for GABARAPL1. Based on previous data reporting an epigenetic regulation of BECN-1 in BC and our data showing a deregulation of DNA methylation in our BC samples (Fig. 2a, and Additional file 1: Figure S1), we hypothesized that the loss of GABARAPL1 expression in BC might be linked to epigenetic modifications. Indeed, DNA methylation dependent gene silencing is frequent in BC and is highly related to BC tumor genotypes (e.g. hypermethylation of ESR1 in ERα negative patients), and may be essential to determine the good treatment [45]. We report here that the GABARAPL1 gene is highly methylated in both–600 (MC1) and +200 (MC2) promoter regions but the 5′-UTR +200 region presents the more significant difference between non tumoral tissues/cells promoter regions and tumoral/cancer cells since this region is poorly methylated in normal tissue and frequently methylated in tumors (Fig. 2). As described in previous studies, a correlation between local DNA methylation and histone modifications is frequently observed in epigenetic-mediated gene silencing [46, 47]. Our results confirm these observations, since we observed that the promoter of GABARAPL1 presented a high level of methylated DNA and deacetylated histone H3 (Fig. 3). On the other hand, DNA methylation seemed poorly involved in GABARAP and GABARAPL2 expression while the inhibition of HDACs by TSA increased H3 acetylation and increased GABARAP and GABARAPL2 expression (Figs. 3 and 4).
Since the control of transcription factor accessibility in the region close to the transcription initiation site is often crucial for gene expression, we wondered whether it might be important for the regulation of GABARAPL1 expression. According to transcription binding site prediction softwares, our data indeed revealed that GABARAPL1 expression is controlled by CREB-1 and that inhibition of epigenetic repressive marks increased CREB-1-recruitment on the GABARAPL1 promoter. Since CREB-1 has been previously involved in the regulation of autophagy, the role of this transcriptional factor may be crucial in this process. Indeed, neuron protection mediated by CREB-1 activation and associated with an increase of BECN-1 and LC3 expression, was observed following rapamycin administration in ischemic neonatal rats [48]. On the opposite, CREB-1 activation together with mTOR inhibitors has also been described to potentiate chemotherapies in renal cancers [49]. Nevertheless, the mechanism by which CREB-1 can regulate autophagy and pro-survival signals in cancer cells will require further studies.
Conclusion
The current use of epigenetic drugs in clinical trials provides new options for personalized adjuvant therapies in cancers. Indeed, HDACi, such as Entinostat or valproic acid efficiently increase ERα expression in ER-BC tumors and considerably improve the efficiency of anti-estrogen signaling therapies [50, 51]. 5-aza-CdR treatment has also been previously reported to increase autophagy by inducing LC3B-II in myeloid cells and myeloid cells resistant to 5-aza-CdR presented an increase of basal autophagy [52]. An increase of GABARAP family protein expression, following 5-aza-CdR/TSA treatment, might at least partly explain this increase of autophagy levels. Indeed, GABARAP/GABARAPL1 overexpression in BC has been described to decrease cell proliferation and tumorigenesis in nude mice [53, 54]. All these data strongly support the idea that autophagy regulation may be a focal point for the design of combined anti-cancer therapies in the future. Mahalingam et al. recently proposed the evaluation of a combination of HDACi (vorinostat) and autophagy inhibitor (hydroxychloroquine) in a phase I study in patients with solid tumors [55]. However, identification of the molecular mechanisms governing gene silencing in autophagy impairment in cancer will definitely help to develop future specific drugs, and decrease the important side effects. Indeed, while DNMTi efficiently restore ERα or BECN-1 expression in methylated tumors, these compounds also reduce global DNA methylation and local methylation [56]. DNMTi also strongly induce metalloproteinase expression in lymphoma and pancreatic cancers suggesting an increase of metastatic potential [57]. Similarly, urokinase, a marker of invasiveness associated with the most aggressive BC and with prostate cancers is increased after DNMTi treatment [58, 59].. In agreement with previous studies on different autophagy related-genes, our work demonstrated for the first time that the GABARAP family genes, and particularly GABARAPL1, are regulated by epigenetic modifications in BC and that epigenetic inhibitors might be used in combination with classical anti-chemotherapeutive drugs for futures anti-cancer therapies.
Abbreviations
- ATG:
-
Autophagy-related protein
- BECN-1:
-
BECLIN-1
- BC:
-
Breast Cancer
- ChIP:
-
Chromatin immunoprecipitation
- CREB-1:
-
cAMP response element binding protein
- DNMT:
-
DNA methyl transferase
- DNMTi:
-
DNMT inhibitor
- ER:
-
Estrogen receptor
- GABARAP:
-
GABAA-receptor associated protein
- GABARAPL1/GEC1:
-
GABARAP like protein 1/Glandular Epithelial Cells 1
- GL1:
-
GABARAPL1
- GABARAPL2/GATE-16:
-
GABARAP like protein 2/golgi-associated ATPase enhancer of 16 kDa
- HDAC:
-
Histone deacetylase
- HDACi:
-
HDAC inhibitor
- HDM:
-
Histone demethylase
- HER:
-
Human epidermal growth factor
- hMOF:
-
Human ortholog of drosophila males absent in the first
- HMT:
-
Histone methyl transferase
- IF:
-
Immunofluorescence
- MAP-LC3:
-
Microtubule associated protein-Light chain 3
- PBS:
-
Phosphate buffer saline
- PBS-T:
-
PBS-triton-×100
- PFA:
-
Paraformaldehyde
- PR:
-
Progesterone receptor
- PVDF:
-
Polyvinylidene difluoride
- SDS:
-
Sodium dodecylsulfate
- TBS-T:
-
Tris buffer saline tween 20
- TSA:
-
Trichostatin A
- 5aza-CdR:
-
5-aza-deoxycytidine
References
Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76.
Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34(7):856–80.
Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6(12):1221–8.
Yu L, Gu C, Zhong D, Shi L, Kong Y, Zhou Z, et al. Induction of autophagy counteracts the anticancer effect of cisplatin in human esophageal cancer cells with acquired drug resistance. Cancer Lett. 2014;355(1):34–45.
Nemos C, Mansuy V, Vernier-Magnin S, Fraichard A, Jouvenot M, Delage-Mourroux R. Expression of gec1/GABARAPL1 versus GABARAP mRNAs in human: predominance of gec1/GABARAPL1 in the central nervous system. Brain Res Mol Brain Res. 2003;119(2):216–9.
Berthier A, Seguin S, Sasco AJ, Bobin JY, De Laroche G, Datchary J, et al. High expression of gabarapl1 is associated with a better outcome for patients with lymph node-positive breast cancer. Br J Cancer. 2010;102(6):1024–31.
Byler S, Goldgar S, Heerboth S, Leary M, Housman G, Moulton K, et al. Genetic and epigenetic aspects of breast cancer progression and therapy. Anticancer Res. 2014;34(3):1071–7.
Vaissiere T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res. 2008;659(1–2):40–8.
Sharma P, Stecklein SR, Kimler BF, Sethi G, Petroff BK, Phillips TA, et al. The prognostic value of promoter methylation in early stage triple negative breast cancer. J Cancer Ther Res. 2014;3(2):1–11.
Guil S, Esteller M. DNA methylomes, histone codes and miRNAs: tying it all together. Int J Biochem Cell Biol. 2009;41(1):87–95.
Fullgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, et al. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500(7463):468–71.
Oh M, Choi IK, Kwon HJ. Inhibition of histone deacetylase1 induces autophagy. Biochem Biophys Res Commun. 2008;369(4):1179–83.
Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29(5):969–80.
Yan J, Seibenhener ML, Calderilla-Barbosa L, Diaz-Meco MT, Moscat J, Jiang J, et al. SQSTM1/p62 interacts with HDAC6 and regulates deacetylase activity. PLoS One. 2013;8(9):e76016.
Bai H, Inoue J, Kawano T, Inazawa J. A transcriptional variant of the LC3A gene is involved in autophagy and frequently inactivated in human cancers. Oncogene. 2012;31(40):4397–408.
Dunwell T, Hesson L, Rauch TA, Wang L, Clark RE, Dallol A, et al. A genome-wide screen identifies frequently methylated genes in haematological and epithelial cancers. Mol Cancer. 2010;9:44.
Li Z, Chen B, Wu Y, Jin F, Xia Y, Liu X. Genetic and epigenetic silencing of the beclin 1 gene in sporadic breast tumors. BMC Cancer. 2010;10:98.
Shukla S, Patric IR, Patil V, Shwetha SD, Hegde AS, Chandramouli BA, et al. Methylation silencing of ULK2, an autophagy gene, is essential for astrocyte transformation and tumor growth. J Biol Chem. 2014;289(32):22306–18.
Swiderek E, Kalas W, Wysokinska E, Pawlak A, Rak J, Strzadala L. The interplay between epigenetic silencing, oncogenic KRas and HIF-1 regulatory pathways in control of BNIP3 expression in human colorectal cancer cells. Biochem Biophys Res Commun. 2013;441(4):707–12.
Pernodet N, Hermetet F, Adami P, Vejux A, Descotes F, Borg C, et al. High expression of QSOX1 reduces tumorogenesis, and is associated with a better outcome for breast cancer patients. Breast Cancer Res. 2012;14(5):R136.
Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86.
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54.
Hervouet E, Demont J, Pecina P, Vojtiskova A, Houstek J, Simonnet H, et al. A new role for the von Hippel-Lindau tumor suppressor protein: stimulation of mitochondrial oxidative phosphorylation complex biogenesis. Carcinogenesis. 2005;26(3):531–9.
Claude-Taupin A, Boyer-Guittaut M, Delage-Mourroux R, Hervouet E. Use of epigenetic modulators as a powerful adjuvant for breast cancer therapies. Methods Mol Biol. 2015;1238:487–509.
Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22(2):246–58.
Tryndyak VP, Kovalchuk O, Pogribny IP. Loss of DNA methylation and histone H4 lysine 20 trimethylation in human breast cancer cells is associated with aberrant expression of DNA methyltransferase 1, Suv4-20 h2 histone methyltransferase and methyl-binding proteins. Cancer Biol Ther. 2006;5(1):65–70.
Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–31.
Seguin-Py S, Lucchi G, Croizier S, Chakrama FZ, Despouy G, Le Grand JN, et al. Identification of HSP90 as a new GABARAPL1 (GEC1)-interacting protein. Biochimie. 2012;94(3):748–58.
Zou CF, Jia L, Jin H, Yao M, Zhao N, Huan J, et al. Re-expression of ARHI (DIRAS3) induces autophagy in breast cancer cells and enhances the inhibitory effect of paclitaxel. BMC Cancer. 2011;11:22.
Mahmoud L, Al-Enezi F, Al-Saif M, Warsy A, Khabar KS, Hitti EG. Sustained stabilization of Interleukin-8 mRNA in human macrophages. RNA Biol. 2014;11(2):124–33.
Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16(1):45–58.
Gao X, Zacharek A, Salkowski A, Grignon DJ, Sakr W, Porter AT, et al. Loss of heterozygosity of the BRCA1 and other loci on chromosome 17q in human prostate cancer. Cancer Res. 1995;55(5):1002–5.
Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Kim SS, et al. Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability. J Pathol. 2009;217(5):702–6.
Kim MS, Jeong EG, Ahn CH, Kim SS, Lee SH, Yoo NJ. Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Hum Pathol. 2008;39(7):1059–63.
Russell SE, Hickey GI, Lowry WS, White P, Atkinson RJ. Allele loss from chromosome 17 in ovarian cancer. Oncogene. 1990;5(10):1581–3.
Saito H, Inazawa J, Saito S, Kasumi F, Koi S, Sagae S, et al. Detailed deletion mapping of chromosome 17q in ovarian and breast cancers: 2-cM region on 17q21.3 often and commonly deleted in tumors. Cancer Res. 1993;53(14):3382–5.
Hervouet E, Hulin P, Vallette FM, Cartron PF. Proximity ligation in situ assay for monitoring the global DNA methylation in cells. BMC Biotechnol. 2011;11:31.
Hervouet E, Lalier L, Debien E, Cheray M, Geairon A, Rogniaux H, et al. Disruption of Dnmt1/PCNA/UHRF1 interactions promotes tumorigenesis from human and mice glial cells. PLoS One. 2010;5(6):e11333.
Hervouet E, Vallette FM, Cartron PF. Impact of the DNA methyltransferases expression on the methylation status of apoptosis-associated genes in glioblastoma multiforme. Cell Death Dis. 2010;1:e8.
He G, Wang Y, Pang X, Zhang B. Inhibition of autophagy induced by TSA sensitizes colon cancer cell to radiation. Tumour Biol. 2014;35(2):1003–11.
Le Grand JN, Chakrama FZ, Seguin-Py S, Fraichard A, Delage-Mourroux R, Jouvenot M, et al. GABARAPL1 (GEC1): original or copycat? Autophagy. 2011;7(10):1098–107.
Cook JL, Re RN, DeHaro DL, Abadie JM, Peters M, Alam J. The trafficking protein GABARAP binds to and enhances plasma membrane expression and function of the angiotensin II type 1 receptor. Circ Res. 2008;102(12):1539–47.
Mansuy V, Boireau W, Fraichard A, Schlick JL, Jouvenot M, Delage-Mourroux R. GEC1, a protein related to GABARAP, interacts with tubulin and GABA (A) receptor. Biochem Biophys Res Commun. 2004;325(2):639–48.
Liu C, Xia Y, Jiang W, Liu Y, Yu L. Low expression of GABARAPL1 is associated with a poor outcome for patients with hepatocellular carcinoma. Oncol Rep. 2014;31(5):2043–8.
Hervouet E, Cartron PF, Jouvenot M, Delage-Mourroux R. Epigenetic regulation of estrogen signaling in breast cancer. Epigenetics. 2013;8(3):237–45.
Ali MW, Cacan E, Liu Y, Pierce JY, Creasman WT, Murph MM, et al. Transcriptional suppression, DNA methylation, and histone deacetylation of the regulator of G-protein signaling 10 (RGS10) gene in ovarian cancer cells. PLoS One. 2013;8(3):e60185.
Cartron PF, Blanquart C, Hervouet E, Gregoire M, Vallette FM. HDAC1-mSin3a-NCOR1, Dnmt3b-HDAC1-Egr1 and Dnmt1-PCNA-UHRF1-G9a regulate the NY-ESO1 gene expression. Mol Oncol. 2012;7(3):452–63.
Carloni S, Girelli S, Scopa C, Buonocore G, Longini M, Balduini W. Activation of autophagy and Akt/CREB signaling play an equivalent role in the neuroprotective effect of rapamycin in neonatal hypoxia-ischemia. Autophagy. 2010;6(3):366–77.
Wang Y, Hu Z, Liu Z, Chen R, Peng H, Guo J, et al. MTOR inhibition attenuates DNA damage and apoptosis through autophagy-mediated suppression of CREB1. Autophagy. 2013;9(12):2069–86.
Fortunati N, Bertino S, Costantino L, De Bortoli M, Compagnone A, Bandino A, et al. Valproic acid restores ER alpha and antiestrogen sensitivity to ER alpha-negative breast cancer cells. Mol Cell Endocrinol. 2010;314(1):17–22.
Sabnis GJ, Goloubeva O, Chumsri S, Nguyen N, Sukumar S, Brodie AM. Functional activation of the estrogen receptor-alpha and aromatase by the HDAC inhibitor entinostat sensitizes ER-negative tumors to letrozole. Cancer Res. 2011;71(5):1893–903.
Cluzeau T, Robert G, Puissant A, Jean-Michel K, Cassuto JP, Raynaud S, et al. Azacitidine-resistant SKM1 myeloid cells are defective for AZA-induced mitochondrial apoptosis and autophagy. Cell Cycle. 2011;10(14):2339–43.
Klebig C, Seitz S, Arnold W, Deutschmann N, Pacyna-Gengelbach M, Scherneck S, et al. Characterization of {gamma}-aminobutyric acid type A receptor-associated protein, a novel tumor suppressor, showing reduced expression in breast cancer. Cancer Res. 2005;65(2):394–400.
Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–6.
Mahalingam D, Mita M, Sarantopoulos J, Wood L, Amaravadi RK, Davis LE, et al. Combined autophagy and HDAC inhibition: a phase I safety, tolerability, pharmacokinetic, and pharmacodynamic analysis of hydroxychloroquine in combination with the HDAC inhibitor vorinostat in patients with advanced solid tumors. Autophagy. 2014;10(8):1403–14.
Cheray M, Pacaud R, Nadaradjane A, Vallette FM, Cartron PF. Specific inhibition of one DNMT1-including complex influences tumor initiation and progression. Clin Epigenetics. 2013;5(1):9.
Sato N, Maehara N, Su GH, Goggins M. Effects of 5-aza-2′-deoxycytidine on matrix metalloproteinase expression and pancreatic cancer cell invasiveness. J Natl Cancer Inst. 2003;95(4):327–30.
Ateeq B, Unterberger A, Szyf M, Rabbani SA. Pharmacological inhibition of DNA methylation induces proinvasive and prometastatic genes in vitro and in vivo. Neoplasia. 2008;10(3):266–78.
Pakneshan P, Xing RH, Rabbani SA. Methylation status of uPA promoter as a molecular mechanism regulating prostate cancer invasion and growth in vitro and in vivo. FASEB J. 2003;17(9):1081–8.
Coulon V, Chebli K, Cavelier P, Blanchard JM. A novel mouse c-fos intronic promoter that responds to CREB and AP-1 is developmentally regulated in vivo. PLoS One. 2010;5(6):e11235.
Arrigo AP, Firdaus WJ, Mellier G, Moulin M, Paul C, Diaz-latoud C, et al. Cytotoxic effects induced by oxidative stress in cultured mammalian cells and protection provided by Hsp27 expression. Methods. 2005;35(2):126–38.
Hervouet E, Debien E, Campion L, Charbord J, Menanteau J, Vallette FM, et al. Folate supplementation limits the aggressiveness of glioma via the remethylation of DNA repeats element and genes governing apoptosis and proliferation. Clin Cancer Res. 2009;15(10):3519–29.
Acknowledgements
We thank Dr. Vincent Coulon (Montpellier, France) for providing the pCDNA3.1-CREB-1 plasmid [60]. This work was supported by the Université de Franche-Comté, and funding from “BQR Jeunes chercheurs” of Université de Franche-Comté, Ministère de l’Enseignement Supérieur de la Recherche (MESR), Région de Franche-Comté 2013 and Ligue Contre le Cancer (#007. Y-2014). ACT was supported by a fellowship from Région de Franche-Comté.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interest
The authors declare that they have no competing interests
Authors’ contributions
EH, ACT, TG, VP carried out the molecular studies. FM and MPA participated in the selection of samples and their validation. EH and MBG wrote the manuscript. AF, PA, GD and RDM helped draft the manuscript. All authors have read and approved the final version of the manuscript
Additional files
Additional file 1: Figure S1.
XIST and GAPDH methylation status in BC. XIST and GAPDH methylation was quantified using methylCollector kit. NT: non tumoral, ER +/−: status of expression of estrogen receptor α, PR+/−: status of expression of progesterone receptor, HER+/−: status of expression of Human epidermal growth factor receptor. White : absence of signal ; black : signal of methylation. (TIFF 77 kb)
Additional file 2: Figure S2.
Effects of 5-aza-CdR/TSA treatment on autophagy and cell proliferation. Increase of both cytosolic LC3-I and autophagosome associated LC3-II forms detected by western blotting using lysates of MCF-7 cells treated with 5-aza-CdR/TSA (antibody anti-LC3: L8918, Sigma-Aldrich). (B) Increase of the number of cells presenting vesicles in GFP-LC3 positive MCF-7 cells transfected with GFP-LC3 and treated with 5-aza-CdR or TSA. (C) A decrease of cell proliferation was observed in MCF-7 cells treated with 5-aza-CdR/TSA using the crystal violet/acid acetic method as previously described [61]. (D) A decrease of clonogenecity was observed in MCF-7 treated with 5-aza-CdR/TSA using the crystal violet method as previously described [62]. (TIFF 122 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Hervouet, E., Claude-Taupin, A., Gauthier, T. et al. The autophagy GABARAPL1 gene is epigenetically regulated in breast cancer models. BMC Cancer 15, 729 (2015). https://doi.org/10.1186/s12885-015-1761-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-015-1761-4