Abstract
Background
While the majority of germline inactivating mutations in BRCA1/2 are small-scale mutations, large genomic rearrangements (LGRs) are also detected in a variable proportion of patients. However, routine genetic methods are incapable of detecting LGRs, and comprehensive genetic testing algorithm is necessary.
Methods
We performed multiplex ligation-dependent probe amplification assay for small-scale mutation negative patients at high-risk for LGR, based on previously published LGR risk criteria. The inclusion criteria for the high-risk subgroup were personal history of 1) early-onset breast cancer (diagnosed at ≤36 years); 2) two breast primaries; 3) breast cancer diagnosed at any age, with ≥1 close blood relatives (includes first-, second-, or third-degree) with breast and/or epithelial ovarian cancer; 4) both breast and epithelial ovarian cancer diagnosed at any age; and 5) epithelial ovarian cancer with ≥1 close blood relatives with breast and/or epithelial ovarian cancer.
Results
Two LGRs were identified. One was a heterozygous deletion of exon 19 and the other was a heterozygous duplication of exon 4–6. The prevalence of LGRs was 7% among Sanger-negative, high-risk patients, and accounted for 13% of all BRCA1 mutations and 2% of all patients. Moreover, LGRs reported in Korean patients, including our 2 newly identified cases, were found exclusively in families with at least one high-risk feature.
Conclusions
Our result suggests that selective LGR screening for Sanger-negative, high-risk patients is necessary for Korean patients.
Similar content being viewed by others
Background
Breast cancer was the second most common cancer in females aged 15–64 in Korea [1] and in 2013, a total of 17,292 incident breast cancer cases were reported in Korea Central Cancer Registry [2]. Breast cancer incidence in Korea has markedly increased in recent years, and the crude incidence of breast cancer in Korea is the highest among Asian countries [3]. Also, the average age at onset of breast cancer is 10 years earlier than Western populations. A younger age at diagnosis suggests that genetic susceptibility genes may be involved in a substantial proportion of breast cancer in Korea.
Recently, genetic counselling and genetic testing of BRCA1 and BRCA2 for the presence of germline inactivating mutations have been increasingly offered to identify individuals at elevated risk of breast and ovarian cancer in Korea. According to a large nationwide prospective Korean Hereditary Breast Cancer (KOHBRA) study, 153 distinct BRCA1/2 mutations have been identified in Korean breast cancer patients with a family history of breast/ovarian cancer resulting in a prevalence of 22.3% [3].
However, until recently, testing of BRCA1 and BRCA2 mutations has been focused on the identification of small-scale mutations (point mutations, small deletions and insertions). Such mutations occur throughout the whole coding sequence and at the splice junctions of both genes. They result in protein truncation, disruption of messenger RNA processing, or amino acid substitutions that have significant impact on protein function and are readily detectable by standard methods of Sanger sequencing of polymerase chain reaction (PCR)-amplified gene segments [4]. Large genomic rearrangement (LGR) of BRCA1 and BRCA2, which is another mechanism of gene inactivation, is responsible for a variable but significant proportion of BRCA mutations [5]. High prevalence of LGRs in BRCA1 have been demonstrated in several populations, including Dutch, Northern Italian, French, and Czech, and in such populations, LGR screening has been advocated as a cost-effective, initial phase screening test [6]. In Korea, reported LGR cases are few, and routine PCR-based genetic testing methods are not capable of detecting LGRs. Therefore, an efficient genetic testing algorithm that incorporates LGR testing is necessary for accurate mutational screening of high-risk patients.
Herein, we performed multiplex ligation-dependent probe amplification (MLPA) for LGR analysis in a subset of small-scale mutation negative patients who were also stratified as high-risk for LGR based on previously published LGR risk criteria from other ethnicities [5, 7]. Here we report the prevalence of the different type of BRCA mutations according to risk stratification, to provide evidence for developing an effective and comprehensive BRCA genetic screening strategy in Korean patients.
Methods
Patients and clinical diagnosis
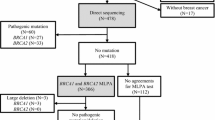
A total of 106 patients at risk for hereditary breast and ovarian cancer (HBOC) and for whom mutation analysis was requested from January 2015 to November 2015 at Seoul St. Mary’s Hospital were included in this study. The family history, past medical history, and tumor pathology of the probands and their family members were detailed by their referring physicians and/or through review of patient’s medical records. All participants gave informed consent, and this study was approved by the Institutional Review Board (IRB)/Ethics Committee of Seoul St. Mary’s Hospital (IRB No.KC15RISI0915).
The referred patients all met the National Comprehensive Cancer Network genetic testing criteria for HBOC syndrome [8] and high-risk subgroup for LGR was defined based on the previously published LGR risk criteria [5, 7, 9]. The inclusion criteria for the high-risk subgroup were personal history of 1) early-onset breast cancer (diagnosed at ≤36 years); 2) two breast cancer primaries; 3) breast cancer diagnosed at any age, with ≥1 close blood relatives (includes first-, second-, or third-degree) with breast and/or epithelial ovarian cancer; 4) both breast and epithelial ovarian cancer diagnosed at any age; and 5) epithelial ovarian cancer with ≥1 close blood relatives with breast and/or epithelial ovarian cancer.
Sanger sequencing
Sanger sequencing was performed in all patients to detect small-scale mutations. Genomic DNA was isolated from the peripheral blood leukocytes, using the QIAmp DNA Mini Kit (Qiagen, Hamburg, Germany). Sanger sequencing was performed as described previously [10]. Exon numbering and DNA sequence variant descriptions are based on NM_007294.3 and NM_000059.3 as reference sequences for BRCA1 and BRCA2. To classify variants, we followed the standards and guidelines of the American College of Medical Genetics and Genomics (ACMG) for the interpretation of sequence variants [11], and all variants were scored and classified into five pathogenicity groups (class 1: benign; class 2: likely benign; class 3: uncertain significance (VUS); class 4: likely pathogenic; class 5; pathogenic).
MLPA analyses
MLPA was performed for all Sanger sequencing-negative patients in the LGR high-risk subgroup. MLPA probe mixes P002 and P045 were used for screening of LGRs in BRCA1 and BRCA2, respectively, and P087 and P077 were used for confirmation, according to the manufacturer’s recommendations (MRC-Holland, Amsterdam, Netherlands). MLPA data were analyzed using Genemarker v1.91 (Softgenetics, State College, PA). Peak heights were normalized and a deletion or duplication was defined as recommended by the manufacturer. Direct sequencing of the probe binding and ligation sites was performed in relevant exons to detect if any polymorphism was located close to the ligation site, which may lead to a false decrease in peak signal [4].
Statistical analyses
Categorical variables were compared using the Chi-square test. Continuous variables were compared using the independent samples t-test or Mann–Whitney-Wilcoxon rank sum tests. MedCalc version 12.1.4 (MedCalc Software, Mariakerke, Belgium) was used and P < 0.05 was considered statistically significant.
Results
Patient risk characteristics
A total of 106 patients at risk of HBOC were enrolled. All were female patients with a mean age of 51 years and mean age at diagnosis was 48 years. Sixty-six patients (62%) were ovarian cancer cases and 40 patients (38%) were breast cancer cases with 7 having both breast and ovarian cancer (Table 1).
When the patient’s risk of HBOC was stratified according to the aforementioned high-risk criteria, 44 patients (35 breast and 9 ovarian cancer patients) were classified as high-risk (Table 1).
BRCA1 and BRCA2 mutation screening
Of the 106 patients, Sanger sequencing identified 11 different BRCA1 small-scale mutations in 13 patients (12%) and 6 BRCA2 small-scale mutations in 9 patients (8%) (Fig. 1). Of the 17 BRCA1/2 small-scale mutations, 1 BRCA1 (c.2345delG) and 1 BRCA2 mutations (c.3445_3446dupAT, n = 3) were novel, and 1 BRCA1 (c.5496_5506delGGTGACCCGAGinsA) and 2 BRCA2 mutations (c.1399A > T, n = 2; c.7480C > T, n = 3) were recurrent. In addition, 13 different BRCA1 VUSs and 22 different BRCA2 VUSs were detected and p.M784V in BRCA2 was the most common VUS (Fig. 1).
Deleterious mutations (red labels) and variants of unknown significance (blue labels) of BRCA1/2 genes found in this study. The allele count of each variant is represented by the number of black square and novel variations are underlined. Red squares represent the detected large genomic rearrangements. Distances between variants and dimensions of each exons are not to scale
MLPA analysis of BRCA1 and BRCA2 in Sanger-negative, high-risk patients revealed 2 previously reported LGRs: a duplication of BRCA1 exon 4–6 and a deletion of BRCA1 exon 19 (Fig. 2). The LGRs consisted 13% of all identified BRCA1 mutations and the prevalence of LGRs identified in this study was 7% in 29 Sanger-negative, high-risk patients and 2% of all enrolled patients.
BRCA1 and BRCA2 mutation prevalence and risk factors
The overall prevalence of BRCA mutations (including both small-scale mutations and LGRs) was 20% of all patients, 30% (12/40) for breast cancer patients, and 18% (12/66) for ovarian cancer patients. Among patients with personal history of breast cancer, BRCA mutations were most frequently detected in those with both breast and ovarian cancer (43%), followed by bilateral breast cancer (38%), and positive family history (37%). Of note, relatively low BRCA mutation frequency was found in patients with early-onset breast cancer (18%). And among patients with personal history of ovarian cancer, BRCA mutations were found in 55% of those patients with the single high-risk criterion of positive family history (Table 2).
Although not statistically significant, each of the high-risk features was more commonly observed in the mutation positive group than in the mutation negative group, except for the high-risk feature of early-onset in breast cancer patients. When the presence of at least one of these high-risk features are compared between the mutation positive group and the mutation negative group, the proportion of patients with at least one high-risk was twice as high in the mutation positive group (71%) than in the mutation negative group (35%), with statistical significance (P = 0.0326).
Risk characteristics of Korean patients with LGR
The two patients identified in this study with LGRs all had at least one high-risk feature (Fig. 3). The index patient with a duplication of BRCA1 exon 4–6 was diagnosed at the age of 67 with invasive serous epithelial ovarian cancer, had 2 close blood relatives with breast and/or epithelial ovarian cancer and also had 2 close blood relatives with cancers other than breast or ovary. The index patient with a deletion of BRCA1 exon 19 had a personal history of early-onset, bilateral, triple-negative breast cancer, had 2 close blood relatives with breast cancer and also had 7 close blood relatives with cancers other than breast or ovary.
Also, when mutation probabilities were retrospectively calculated in previously reported Korean LGR probands [12–14] including our 2 newly identified cases by BRCAPRO [15–17] and Korean hereditary breast cancer study BRCA risk calculator (KOHCal) [18], 67% of patients presented with BRCAPRO mutation probability >10% and 100% of patients presented with KOHCal mutation probability of >10%. Moreover, all reported LGRs in Korea were detected in patients with at least one high-risk feature (Table 3).
Discussion
In this report of 106 consecutive Korean patients at risk for HBOC from a single center cohort, we identified 2 LGRs in Sanger-negative, high-risk patients. Also, 11 BRCA1 and 6 BRCA2 small-scale mutations were identified and our report extends the spectrum of BRCA mutations by detecting two novel frame-shift mutations in BRCA1/2. The overall prevalence of BRCA mutations was 20% for all patients, 30% for breast cancer, and 18% for ovarian cancer patients.
The frequency of mutations was related to the type as well as the number of risk factors. Strong predictors of the likelihood of carrying a BRCA mutation in patients with personal history of breast cancer were the occurrence of both breast and ovarian cancer (43%), bilateral breast cancer (38%), and positive family history (37%), and the presence of positive family history (55%) in patients with personal history of ovarian cancer. However, there was no difference between the proportion of early-onset breast cancer patients between the mutation positive group and the mutation negative group. Furthermore, all of the BRCA mutation-positive patients with early-onset breast cancer, had multiple risk factors other than early-onset. This is in agreement with earlier reports that in Korean, non-familial, early-onset breast cancer patients without other risk factors, the prevalence of BRCA mutation is low [3, 18].
The most important finding of this study is that selective screening of high-risk, Sanger-negative Korean patients for LGRs using MLPA analysis, identified 2 patients with LGRs in BRCA1. The LGRs comprised 2% of all enrolled patients but accounted for 7% of Sanger-negative, high-risk patients and 12% of all identified BRCA mutations in high-risk patients. The frequency of LGR varies considerably among populations, with LGRs accounting for one third of BRCA1 mutations in northern Italy, and 27–36% of BRCA1 mutations in Netherlands, but less common in other populations [9]. LGRs in BRCA are considered to be rare in Korea, with a reported frequency of 0.45% in familial breast cancer patients [12] and 2.1% in Sanger-negative, familial breast cancer patients [14]. And, only 5 LGR cases have been reported so far in Korea [3]. However, the characterization of risk characteristics of LGRs in any given population allows a more efficient and cost-effective mutational screening approach. Our results suggest that risk stratification and selectively screening Sanger-negative, high-risk patients for LGRs is an effective screening strategy for LGR detection in Korean patients. And with emerging therapies, such as poly ADP ribose polymerase inhibitors in combination with conventional treatment [19], a comprehensive genetic evaluation strategy encompassing LGRs as well as small-scale mutations has become even more critical.
This study has certain limitations. We did not perform MLPA for all Sanger-negative patients, but only in high-risk patients, and the number of LGR cases is limited. And since, Sanger-negative, non-high-risk patients were not included in the MLPA analysis, the true frequency of LGRs may be greater than the reported 2% of all enrolled patients. However, previous reports have indicated that patients harboring LGR appears to be at the highest end of the range of risks associated with BRCA mutation [5, 9], and the probability of an LGR among non-high-risk patients can be regarded as significantly low. Whereas the present study is limited by small sample size and was not designed to provide a comprehensive survey of the frequency of LGRs, our data suggest that in high-risk families with a negative Sanger sequencing result, LGRs represent a significant source of BRCA mutations.
Conclusions
In summary, we have applied a simple LGR risk criteria, and have shown that screening Sanger-negative high-risk patients for LGR is an effective and necessary genetic testing strategy in Korean patients. Based on the results of this study, risk stratification of at risk patients for HBOC and selective screening for LGRs in BRCA1 is recommended for Korean patients.
Abbreviations
- HBOC:
-
Hereditary breast and ovarian cancer
- IRB:
-
Institutional review board
- KOHBRA:
-
Korean hereditary breast cancer (study)
- KOHCal:
-
Korean hereditary breast cancer study BRCA risk calculator
- LGR:
-
Large genomic rearrangement
- MLPA:
-
Multiplex ligation-dependent probe amplification
- PCR:
-
Polymerase chain reaction
References
Jung KW, Won YJ, Kong HJ, Oh CM, Cho H, Lee DH, Lee KH. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2012. Cancer Res Treat. 2015;47(2):127–41.
Statistics Korea. Korean Statistical Information Service. http://kosis.kr. Accessed 9 May 2016.
Kang E, Seong MW, Park SK, Lee JW, Lee J, Kim LS, Lee JE, Kim SY, Jeong J, Han SA, et al. The prevalence and spectrum of BRCA1 and BRCA2 mutations in Korean population: recent update of the Korean Hereditary Breast Cancer (KOHBRA) study. Breast Cancer Res Treat. 2015;151(1):157–68.
Ewald IP, Ribeiro PL, Palmero EI, Cossio SL, Giugliani R, Ashton-Prolla P. Genomic rearrangements in BRCA1 and BRCA2: A literature review. Genet Mole Biol. 2009;32(3):437–46.
Woodward AM, Davis TA, Silva AG, Kirk JA, Leary JA. Large genomic rearrangements of both BRCA2 and BRCA1 are a feature of the inherited breast/ovarian cancer phenotype in selected families. J Med Genet. 2005;42(5):e31.
Sluiter MD, van Rensburg EJ. Large genomic rearrangements of the BRCA1 and BRCA2 genes: review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res Treat. 2011;125(2):325–49.
Engert S, Wappenschmidt B, Betz B, Kast K, Kutsche M, Hellebrand H, Goecke TO, Kiechle M, Niederacher D, Schmutzler RK, et al. MLPA screening in the BRCA1 gene from 1,506 German hereditary breast cancer cases: novel deletions, frequent involvement of exon 17, and occurrence in single early-onset cases. Hum Mutat. 2008;29(7):948–58.
NCCN Guidelines® Genetic/Familial High-Risk Assessment: Breast and Ovarian. National Comprehensive Cancer Network. http://www.nccn.org/. Accessed 9 May 2016.
James PA, Sawyer S, Boyle S, Young MA, Kovalenko S, Doherty R, McKinley J, Alsop K, Beshay V, Harris M, et al. Large genomic rearrangements in the familial breast and ovarian cancer gene BRCA1 are associated with an increased frequency of high risk features. Fam Cancer. 2015;14(2):287–95.
Park J, Jang W, Chae H, Kim Y, Chi HY, Kim M. Comparison of Targeted Next-Generation and Sanger Sequencing for the BRCA1 and BRCA2 Mutation Screening. Ann Lab Med. 2016;36(2):197–201.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24.
Cho JY, Cho DY, Ahn SH, Choi SY, Shin I, Park HG, Lee JW, Kim HJ, Yu JH, Ko BS, et al. Large genomic rearrangement of BRCA1 and BRCA2 genes in familial breast cancer patients in Korea. Fam Cancer. 2014;13(2):205–11.
Seong MW, Cho SI, Noh DY, Han W, Kim SW, Park CM, Park HW, Kim SY, Kim JY, Park SS. Low contribution of BRCA1/2 genomic rearrangement to high-risk breast cancer in the Korean population. Fam Cancer. 2009;8(4):505–8.
Seong MW, Cho SI, Kim KH, Chung IY, Kang E, Lee JW, Park SK, Lee MH, Choi DH, Yom CK, et al. A multi-institutional study of the prevalence of BRCA1 and BRCA2 large genomic rearrangements in familial breast cancer patients. BMC Cancer. 2014;14:645.
Berry DA, Iversen Jr ES, Gudbjartsson DF, Hiller EH, Garber JE, Peshkin BN, Lerman C, Watson P, Lynch HT, Hilsenbeck SG, et al. BRCAPRO validation, sensitivity of genetic testing of BRCA1/BRCA2, and prevalence of other breast cancer susceptibility genes. J Clin Oncol. 2002;20(11):2701–12.
Gadzicki D, Evans DG, Harris H, Julian-Reynier C, Nippert I, Schmidtke J, Tibben A, van Asperen CJ, Schlegelberger B. Genetic testing for familial/hereditary breast cancer-comparison of guidelines and recommendations from the UK, France, the Netherlands and Germany. J Community Genet. 2011;2(2):53–69.
Parmigiani G, Berry D, Aguilar O. Determining carrier probabilities for breast cancer-susceptibility genes BRCA1 and BRCA2. Am J Hum Genet. 1998;62(1):145–58.
Kang E, Park SK, Lee JW, Kim Z, Noh WC, Jung Y, Yang JH, Jung SH, Kim SW. KOHBRA BRCA risk calculator (KOHCal): a model for predicting BRCA1 and BRCA2 mutations in Korean breast cancer patients. J Hum Genet. 2016;61(5):365–71.
Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–21.
Acknowledgements
We are grateful to the patients and their treating physicians who participated in this study, and also acknowledge the support of Catholic Genetic Laboratory Center for carrying out this study.
Funding
No funding to declare.
Availability of data and materials
“The raw datasets are not publicly available owing to the study’s data access policies. However, analyses in the dataset can be requested by contacting the corresponding author.”
Authors’ contributions
DHK and HC wrote the manuscript, coordinated the research. KHL, SYH, BJC and BJS obtained informed consent and collected clinical data and blood samples from patients. DHK, HC, IJ, JY, HL, WJ, JP, GDL and DSJ performed the genetic studies. MK and YK designed the study and revised the manuscript for important intellectual content. All authors have read and approved the final manuscript and its submission for publication.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
All participants gave informed consent, and this study was approved by the Institutional Review Board (IRB)/Ethics Committee of Seoul St. Mary’s Hospital (IRB No.KC15RISI0915).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kim, DH., Chae, H., Jo, I. et al. Identification of large genomic rearrangement of BRCA1/2 in high risk patients in Korea. BMC Med Genet 18, 38 (2017). https://doi.org/10.1186/s12881-017-0398-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-017-0398-3