Abstract
Background
Large genomic rearrangements (LGRs) in the BRCA1/2 genes are frequently observed in breast cancer patients who are negative for BRCA1/2 small mutations. Here, we examined 221 familial breast cancer patients from 37 hospitals to estimate the contribution of LGRs, in a nationwide context, to the development of breast cancer.
Methods
Direct sequencing or mutation scanning followed by direct sequencing was performed to screen small mutations. BRCA1/2 small mutation-negative patients were screened for the presence of LGRs using a multiple ligation-dependent probe amplification (MLPA) assay.
Results
Using a combined strategy to detect the presence of small mutations and LGRs, we identified BRCA1/2 small mutations in 78 (35.3%) out of 221 familial breast cancer patients and BRCA1 LGRs in 3 (2.1%) out of 143 BRCA1/2 small mutation-negative patients: the deletion of exons 11–13, the deletion of exons 13–15, and whole gene deletion of exons 1-24. The novel deletion of exons 11–13 is thought to result from a non-homologous recombination event mediated by a microhomology sequence comprised of 3 or 4 base pairs: c.3416_4357 + 1863delins187 (NG_005905.2: g.33369_44944delins187).
Conclusions
In this study, we showed that LGRs were found in 3.7% (3/81) of the patients who had mutations in BRCA1 or BRCA2, and 7.5% (3/40) of patients with mutations in BRCA1. This suggests that the contribution of LGRs to familial breast cancer in this population might be comparable to that in other ethnic populations. Given these findings, an MLPA to screen for mutations in the BRCA1 gene is recommended as an initial screening test in highly selective settings.
Similar content being viewed by others
Background
BRCA1 and BRCA2 are the two major genes that contribute to the development of hereditary breast cancer. Mutations in these two genes are observed in 15–20% of hereditary breast cancer cases but less than 5% of overall breast cancer cases [1, 2]. Most BRCA1 or BRCA2 gene aberrations are small mutations involving a single or multiple changes in nucleotide sequence, but large genomic rearrangements (LGRs) have also been reported in patients who are negative for BRCA1 or BRCA2 sequence variations. The prevalence of LGRs varies according to the ethnicity and selection criteria of the study population [3, 4]. In certain studies conducted in the Netherlands and Italy, it was shown that in breast and/or ovarian cancer patients who tested negative for BRCA1 or BRCA2 small mutations, about 20% carried an LGR in 1 of these 2 genes [5, 6], but many other studies reported a prevalence of under 10% [7, 8]. Although the prevalence of LGRs is expected to be low in the population in this study compared to other ethnic populations, the findings reported here are based on a limited study and the nationwide prevalence is not known yet in this population [9, 10].
Here, we analyzed 221 familial breast and/or ovarian cancer patients from 37 different hospitals throughout Korea to estimate the contribution of LGRs to the development of breast cancer in a nationwide context.
Methods
Subjects
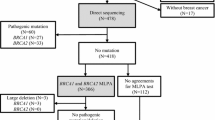
Patients who fulfilled the following criteria were selected for this study between February 2006 and November 2011: female patients with breast cancer diagnosed at any age, patients with a family history of 2 or more breast cancer cases or 1 or more ovarian cancer cases. In total, 221 patients were enrolled from 37 hospitals throughout the country. The mutational status of the BRCA1 and BRCA2 genes was determined for all patients by using the following approaches: direct sequencing (155 patients), or mutation scanning including fluorescence-based conformation-sensitive gel electrophoresis (F-CSGE) and denaturing high-performance liquid chromatography (dHPLC) followed by direct sequencing (66 patients) (Figure 1). In the latter approach, direct sequencing was performed when a patient was found to be positive using the F-CSGE or dHPLC (14 patients) methods; direct sequencing was otherwise carried out for the entire gene (52 patients). Excluding the 78 patients who had mutations identified by any of the above approaches, the remaining 143 patients were screened for LGRs. All patients received genetic counseling and BRCA genetic testing was performed after obtaining informed consent. This study was approved by the institutional review board of Seoul National University Bundang Hospital.
Mutation screening
Genomic DNA was extracted from peripheral blood samples using Gentra PureGene DNA Isolation Kits (Gentra Systems, Inc. Minneapolis, MN). F-CSGE and dHPLC were performed as previously described [11]. PCR was performed using primers the flanked the splice junctions of the coding exons [12]. Amplified products were sequenced using an ABI 3730 analyzer (Applied Biosystems, Foster City, CA), with Bigdye Terminator v3.1 Cycle Sequencing Kits. The sequences were analyzed using SeqScape software (Applied Biosystems, Foster City, CA) and Mutation Surveyor (Softgenetics, State College, PA).
Multiplex ligation-dependent probe amplification (MLPA)
BRCA1 and BRCA2 LGRs were screened using MLPA. MLPA was performed using the SALSA P002/P002B BRCA1 Kit (MRC Holland, Amsterdam, Holland) for BRCA1 and P045/P045B BRCA2 Kit (MRC Holland, Amsterdam, Holland) for BRCA2. PCR products were analyzed using an ABI 3100 analyzer with Genemarker v1.51 (Softgenetics, State College, PA). Peak heights were normalized, and a deletion or duplication was identified when the normalized peak ratio value was below 0.75 or above 1.30.
Characterization of BRCA1LGR
Long-range PCR was performed with the primers 1 F-5′-GGAACTAACCAAACGGAGCA-3′ and 1R-5′-AGGATTGCTTGAGCCTGAAA-3′ using genomic DNA template isolated from patient samples showing a large deletion in exons 11–13. The PCR cycling conditions were as follows: an initial denaturation at 94°C for 2 min; 10 cycles at 94°C for 15 s, 65°C for 30 s, and 68°C for 7 min; 25 cycles at 94°C for 15 s and 65°C for 30 s; a final extension at 68°C for 7 min (increase of 20 s per cycle). Amplified products were sequenced with additional sequencing primers (4 F-5′-AACCACAGTCGGGAAACAAG-3′, 5 F-5′- TAGGGGTTTTGCAACCTGAG-3′) using an ABI 3730 analyzer (Applied Biosystems, Foster City, CA) and analyzed using SeqScape software (Applied Biosystems, Foster City, CA) and Mutation Surveyor (Softgenetics, State College, PA). The deletion was described using the nomenclature used for sequence variations as recommended by the Human Genome Variation Study (http://www.hgvs.org/mutnomen); NM_007294.3 was used as the coding DNA reference sequence.
Statistical analysis
The comparisons between the groups were performed using the results from Mann-Whitney U tests. Statistical data was analyzed using SPSS version 16.0 (SPSS, Inc.).
Results
We identified BRCA1/2 small mutations in 78 (35.3%) out of 221 familial breast cancer patients using direct sequencing, or mutation scanning including F-CSGE and dHPLC followed by direct sequencing, and identified BRCA1 LGRs in 3 (2.1%) out of 143 BRCA1/2 small mutation-negative patients using MLPA (Figure 1). In total, the prevalence of BRCA1/2 mutations was 36.7% (81 of 221) in this study. The prevalence of BRCA1 mutations (49.4%, 40/81) was slightly lower than that of BRCA2 mutations (50.6%, 41/81). The three different BRCA1 LGRs identified in the study were as follows: the deletion of exons 11–13, the deletion of exons 13–15 and a whole gene deletion of exons 1-24 (Table 1; Figure 2).
Patient P1, who was diagnosed with carcinoma in situ at the age of 35, carries a BRCA1 gene with deletions of exons 11–13. She had two second-degree relatives with breast cancer. An immunohistochemistry (IHC) study revealed that her tumor was negative for ER, PR, and HER2. The deletion of exons 11–13 was characterized using long-range PCR and subsequent direct sequencing. The patient was identified as having a novel, 11,389 bp-sized deletion that ranged from the base pair at position 681 of the 3′ end of exon 11 to intron 13: c.3416_4357 + 1863delins187 (NG_005905.2: g.33369_44944delins187) (Figure 3). The 5′ breakpoint was not correlated with any Alu element, but the 3′ breakpoint was located within an AluJo element in intron 13. Interestingly, a partial sequence (187 bp) from intron 12 was inserted at the deletion site. Both ends of the inserted sequences showed a 3 base pair (GAA) or 4 base pair (TGTG) microhomology with the 5′ and 3′ breakpoints, respectively.
A detailed characterization of the deletion of exons 11–13 in the BRCA1 gene identified in patient P1. A) Long-range PCR results showing an approximately 11 kb-sized deletion in the BRCA1 gene; B) DNA sequence analysis showing that the deletion in the BRCA1 gene in this case corresponds to NM_007294.3: c.3416_c.4357 + 1863delins187 [NG_005905.2: g.33369_44944delins187]. The 5′ and 3′ breakpoints in the gene are located within exon 11 and intron 13, respectively, with a partial insertion of intron 12 sequence at the breakpoint (c.4185 + 2603_2789). Short common sequences (microhomology) are shown shaded in light gray between the rearranged sequences at each breakpoint. MX and MIV: molecular weight ladders; P1: patient 1; C: healthy control.
Patient P2, who was diagnosed with stage II invasive ductal carcinoma at the age of 35, carries a BRCA1 gene lacking exons 13–15 . ER and PR were positive on IHC staining. The patient has 1 first-degree relative with ovarian cancer and 1 second-degree relative with breast cancer. The precise characterization of the deletion breakpoint in this patient has been previously reported; this deletion may be the result of Alu-mediated homologous recombination events [9].
Patient P3 was diagnosed with stage II invasive ductal carcinoma at age 36 and does not have a copy of the BRCA1 gene. The patient has 4 second-degree relatives with breast cancer. An IHC study revealed that PR was positive, and ER and HER2 were negative. The deletion breakpoint is uncharacterized in this patient.
The mean age at diagnosis of the 3 patients with BRCA1 LGRs was younger than that of patients with non-LGR mutations in BRCA1/2 (35.3 vs. 40.7) or BRCA1 (35.3 vs. 36.8), but these differences were not statistically significant.
Discussion
To the best of our knowledge, this is the first nationwide study reporting a screen for BRCA1/2 LGRs in familial breast and/or ovarian cancer patients in Korea. In this study, 3 LGRs were identified in 3 out of 143 BRCA1 and BRCA2 small mutation-negative patients. Each patient had an LGR unique to the family. Furthermore, LGRs were found only in the BRCA1 gene. The most common mechanism of LGRs found in BRCA1/2 is known as the Alu-mediated unequal homologous recombination, followed by non-homologous events such as Alu/non-Alu or non-Alu/non-Alu, and a recombination event between the BRCA1 gene and a pseudogene [4]. This mechanism occurs because of the high Alu density (41.5%) in the BRCA1 gene, which is 4-fold higher than that in the human genome and 2-fold that observed in the BRCA2 gene [13, 14]. Between 2 fully characterized LGRs in our study, 1 LGR (a deletion of exons 13–15) might occur because of Alu-mediated homologous recombination, and the other (a deletion of exons 11–13) because of non-homologous recombination events that are mediated by microhomology. Non-homologous recombination events frequently result in deletions and short insertions at the site of the deletion [15]. Interestingly, our case showed a relatively long insertion of 187 bp at the deletion site.
The prevalence of BRCA1 LGRs ranges from approximately 6%–27% of all mutations detected in the BRCA1 gene; BRCA2 LGRs play a minimal role in breast cancer [4]. In non-Ashkenazi Jewish families in the United States, BRCA1 and BRCA2 LGRs comprise 18% (8 of 44) of all identified mutations; 29% (8 of 28) and 6% (1 of 16) of these mutations occur in the BRCA1 and BRCA2 gene, respectively [16]. Among Asians from Singapore, LGRs account for 10% (4 of 40) of all mutations and 10.5% (2 of 19) and 9.5% (2/21) in the BRCA1 and BRCA2 gene, respectively [17]. So far, there have only been 2 reports describing BRCA1 LGRs in this population [9, 10]. In the former study, LGRs constituted 2.5% of all BRCA1/2 mutations and 6.2% of BRCA1 mutations and LGRs were rarely detected; LGRs were found only in 1.2% of all BRCA1/2 gene mutations and 2.3% of BRCA1 gene mutations. Our results demonstrating that 3.7% (3/81) of patients who have mutations in BRCA1/2 and 7.5% (3/40) of patients who have a mutation in the BRCA1 gene were higher than those reported previously.
Conclusions
This nationwide study suggests that the contribution of LGRs in the development of familial breast cancer in the Korean population might be comparable to other ethnic populations and a MLPA screening for mutations in the BRCA1 gene is recommended as an initial screening test in highly selective settings.
Abbreviations
- DHPLC:
-
Denaturing high-performance liquid chromatography
- SSCP:
-
Single strand conformational polymorphism
- MLPA:
-
Multiple ligation-dependent probe amplification
- LGR:
-
Large genomic rearrangement
- F-CSGE:
-
Fluorescence-based conformation-sensitive gel electrophoresis.
References
Hall MJ, Reid JE, Burbidge LA, Pruss D, Deffenbaugh AM, Frye C, Wenstrup RJ, Ward BE, Scholl TA, Noll WW: BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer. 2009, 115 (10): 2222-2233. 10.1002/cncr.24200.
Nathanson KL, Wooster R, Weber BL: Breast cancer genetics: what we know and what we need. Nat Med. 2001, 7 (5): 552-556. 10.1038/87876.
Judkins T, Rosenthal E, Arnell C, Burbidge LA, Geary W, Barrus T, Schoenberger J, Trost J, Wenstrup RJ, Roa BB: Clinical significance of large rearrangements in BRCA1 and BRCA2. Cancer. 2012, 118 (21): 5210-5216. 10.1002/cncr.27556.
Sluiter MD, van Rensburg EJ: Large genomic rearrangements of the BRCA1 and BRCA2 genes: review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res Treat. 2011, 125 (2): 325-349. 10.1007/s10549-010-0817-z.
Montagna M, Dalla Palma M, Menin C, Agata S, De Nicolo A, Chieco-Bianchi L, D’Andrea E: Genomic rearrangements account for more than one-third of the BRCA1 mutations in northern Italian breast/ovarian cancer families. Hum Mol Genet. 2003, 12 (9): 1055-1061. 10.1093/hmg/ddg120.
Hogervorst FB, Nederlof PM, Gille JJ, McElgunn CJ, Grippeling M, Pruntel R, Regnerus R, van Welsem T, van Spaendonk R, Menko FH, Kluijt I, Dommering C, Verhoef S, Schouten JP, van’t Veer LJ, Pals G: Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res. 2003, 63 (7): 1449-1453.
Walsh T, Casadei S, Coats KH, Swisher E, Stray SM, Higgins J, Roach KC, Mandell J, Lee MK, Ciernikova S, Foretova L, Soucek P, King MC: Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA. 2006, 295 (12): 1379-1388. 10.1001/jama.295.12.1379.
Mazoyer S: Genomic rearrangements in the BRCA1 and BRCA2 genes. Hum Mutat. 2005, 25 (5): 415-422. 10.1002/humu.20169.
Seong MW, Cho SI, Noh DY, Han W, Kim SW, Park CM, Park HW, Kim SY, Kim JY, Park SS: Low contribution of BRCA1/2 genomic rearrangement to high-risk breast cancer in the Korean population. Fam Cancer. 2009, 8 (4): 505-508. 10.1007/s10689-009-9279-z.
Cho JY, Cho DY, Ahn SH, Choi SY, Shin I, Park HG, Lee JW, Kim HJ, Yu JH, Ko BS, Ku BK, Son BH: Large genomic rearrangement of BRCA1 and BRCA2 genes in familial breast cancer patients in Korea. Fam Cancer. 2014, 13 (2): 205-211. 10.1007/s10689-014-9704-9.
Kim H, Cho DY, Choi DH, Choi SY, Shin I, Park W, Huh SJ, Han SH, Lee MH, Ahn SH, Son BH, Kim SW, Korean Breast Cancer Study Group, Haffty BG: Characteristics and spectrum of BRCA1 and BRCA2 mutations in 3,922 Korean patients with breast and ovarian cancer. Breast Cancer Res Treat. 2012, 134 (3): 1315-1326. 10.1007/s10549-012-2159-5.
Seong MW, Cho SI, Noh DY, Han WS, Kim SW, Park CM, Park HY, Kim SY, Kim JY, Park SS: Comprehensive mutational analysis of BRCA1/BRCA2 for Korean breast cancer patients and evidence of a founder mutation. Clin Genet. 2009, 76 (2): 152-160. 10.1111/j.1399-0004.2009.01202.x.
Ewald IP, Ribeiro PL, Palmero EI, Cossio SL, Giugliani R, Ashton-Prolla P: Genomic rearrangements in BRCA1 and BRCA2: a literature review. Genet Mol Biol. 2009, 32 (3): 437-446. 10.1590/S1415-47572009005000049.
Zhang W, Edwards A, Fan W, Deininger P, Zhang K: Alu distribution and mutation types of cancer genes. BMC Genomics. 2011, 12: 157-10.1186/1471-2164-12-157.
Hastings PJ, Lupski JR, Rosenberg SM, Ira G: Mechanisms of change in gene copy number. Nat Rev Genet. 2009, 10 (8): 551-564. 10.1038/nrg2593.
Palma MD, Domchek SM, Stopfer J, Erlichman J, Siegfried JD, Tigges-Cardwell J, Mason BA, Rebbeck TR, Nathanson KL: The relative contribution of point mutations and genomic rearrangements in BRCA1 and BRCA2 in high-risk breast cancer families. Cancer Res. 2008, 68 (17): 7006-7014. 10.1158/0008-5472.CAN-08-0599.
Lim YK, Lau PT, Ali AB, Lee SC, Wong JE, Putti TC, Sng JH: Identification of novel BRCA large genomic rearrangements in Singapore Asian breast and ovarian patients with cancer. Clin Genet. 2007, 71 (4): 331-342. 10.1111/j.1399-0004.2007.00773.x.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/14/645/prepub
Acknowledgements
This study was supported by grant no. 02-2009-032 from the SNUH Research Fund and by a grant from the National R & D Program for Cancer Control, Ministry for Health, Welfare, and Family Affairs, Republic of Korea (No. 1020350).
We thank all the participants and investigators of the KOHBRA Study: Beom Seok Kwak, Byeong-Woo Park, Byung Ho Son, Byung-In Moon, Cha Kyong Yom, Chan Heun Park, Chan Seok Yoon, Chang Hyun Lee, Dae Sung Yoon, Dong-Young Noh, Doo Ho Choi, Eundeok Chang, Eun-Kyu Kim, Eunyoung Kang, Hae Kyung Lee, Hai-Lin Park, Hyde Lee, Hyeong-Gon Moon, Hyun-Ah Kim, Il-Kyun Lee, Jeong Eon Lee, Jihyoun Lee, Jong Won Lee, Jong-Han Yu, Joon Jeong, Jung Han Yoon, Jung-Hyun Yang, Keumhee Kwak, Ki-Tae Hwang, Ku Sang Kim, Lee Su Kim, Min Hee Hur, Min Ho Park, Min Hyuk Lee, Myung Chul Chang, Nam Sun Paik, Sang Ah Han, Sang Seol Jung, Sang Uk Woo, Se Jeong Oh, Sehwan Han, Sei Joong Kim, Sei-Hyun Ahn, Seok-Jin Nam, Seung Sang Ko, Sung Hoo Jung, Sung Soo Kang, Sung Yong Kim, Sung-Won Kim, Tae Hyun Kim, Tae Wan Won, Tae Woo Kang, Wonshik Han, Woo-Chul Noh, Yong Lai Park, Yongsik Jung, Young Jin Suh, Young Tae Bae, Young Up Cho, Young-Ik Hong, Sue K. Park, Yoon Joo Jung, Su Yun Choi, Young Bum Yoo, Soo-Jung Lee.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SMW and KSW were involved in study conception and design. All authors were involved in data acquisition, analysis and interpretation for this study. SMW wrote the manuscript. All authors have read, revised critically for intellectual content and approved the final version of the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Seong, MW., Cho, S.I., Kim, K.H. et al. A multi-institutional study of the prevalence of BRCA1 and BRCA2 large genomic rearrangements in familial breast cancer patients. BMC Cancer 14, 645 (2014). https://doi.org/10.1186/1471-2407-14-645
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2407-14-645