Abstract
Background
Understanding the relationship between serotype epidemiology and antimicrobial susceptibility of Streptococcus pneumoniae is essential for the effective introduction of pneumococcal conjugate vaccines (PCVs) and control of antimicrobial-resistant pneumococci.
Methods
We conducted a community-based study in Nha Trang, central Vietnam, to clarify the serotype distribution and pattern of S. pneumoniae antimicrobial susceptibility in children under 5 years of age and to identify risk factors for carrying antimicrobial-resistant strains. Nasopharyngeal swabs collected from children with acute respiratory infections (ARIs) hospitalized between April 7, 2008, and March 30, 2009, and from healthy children randomly selected in July 2008 were subjected to bacterial culture. Minimum inhibitory concentrations (MICs) against S. pneumoniae were determined, and multiplex-polymerase chain reaction (PCR) serotyping assays were performed. Logistic regression was applied to identify risk factors.
Results
We collected 883 samples from 331 healthy children and 552 ARI cases; S. pneumoniae was isolated from 95 (28.7%) healthy children and 202 (36.6%) ARI cases. Age and daycare attendance were significantly associated with pneumococcal carriage. In total, 18.0, 25.8 and 75.6% of the isolates had high MICs for penicillin (≥4 μg/ml), cefotaxime (≥2 μg/ml) and meropenem (≥0.5 μg/ml), respectively. The presence of pneumococci non-susceptible to multiple beta-lactams was significantly associated with serotype 19F (Odds Ratio: 4.23) and daycare attendance (Odds Ratio: 2.56) but not ARIs, age or prior antimicrobial use. The majority of isolates non-susceptible to multiple beta-lactams (90%) were PCV13 vaccine serotypes.
Conclusions
S. pneumoniae serotype 19F isolates non-susceptible to multiple beta-lactams are widely prevalent among Vietnamese children. Vaccine introduction is expected to significantly increase drug susceptibility.
Similar content being viewed by others
Background
Streptococcus pneumoniae causes an enormous global disease burden, especially in developing countries [1]. Approximately 0.4 million deaths from pneumococcal pneumonia in children younger than 5 years of age are estimated to occur annually, of which approximately 80% occur in Africa and southeast Asia [1]. In the developed world, where the pneumococcal conjugate vaccine (PCV) for infants has been introduced, both the incidence of invasive pneumococcal disease (IPD) and the hospitalization rate for IPD have decreased [2, 3]. In addition to IPD, reductions in all-cause pneumonia, pneumococcal pneumonia hospitalizations and inpatient mortality have been observed [4, 5].
Several previous epidemiological studies in Vietnam have shown that antimicrobial resistance, including penicillin-resistant S. pneumoniae (PRSP), is quite common among both clinical and carriage isolates [6,7,8,9,10]. Multidrug-resistant, globally circulating clones (Spain23F-1 and Taiwan19F-14) and their related strains are primarily responsible for resistance [7, 9]. Serogroups/serotypes 19, 23, 14 and 6 are predominant in Vietnam [6, 7]. However, no previous studies using community-based surveys have demonstrated the risk of carrying antimicrobial-resistant S. pneumoniae among healthy children or increased resistance to a wide range of beta-lactam antimicrobials, including carbapenems.
The introduction of PCV in the USA reduced vaccine serotype non-susceptible S. pneumoniae carriage and disease [11, 12]. However, compensatory increases in less susceptible non-vaccine types have been subsequently observed [13, 14]. A recent double-blind study confirmed that the 13-valent pneumococcal conjugate vaccine (PCV-13) was superior to the 7-valent pneumococcal conjugate vaccine (PCV-7) in reducing the non-susceptible S. pneumoniae carriage [15]. Understanding the serotype distribution of colonized S. pneumoniae and how the distribution relates to antimicrobial susceptibility is essential for the effective introduction of PCV and the control of drug resistance.
The main objectives of the present study are to clarify the serotype distribution and pattern of antimicrobial susceptibility of S. pneumoniae isolates colonizing children under 5 years of age in Nha Trang, central Vietnam, and to determine the proportion of vaccine serotypes among the resistant strains.
Methods
Study setting
A community-based S. pneumoniae colonization study was conducted in Nha Trang city in central Vietnam. Nha Trang, the capital city of Khanh Hoa province, is an optimal geographic location for population-based surveillance because the eastern side of the city is located on the sea, and the other borders are surrounded by mountains [16].
Nasopharyngeal swab samples were consecutively collected from children under 5 years of age with acute respiratory infections (ARIs) who were admitted to the pediatric ward at Khanh Hoa General Hospital between April 7, 2008, and March 30, 2009. This hospital has a capacity of 1000 beds and is the only hospital providing in-patient care in the city. Nasopharyngeal samples were collected at the time of hospital admission before starting antimicrobial treatment in the pediatric ward. The inclusion criteria for these ARI cases were described elsewhere [17]. To obtain population-representative nasopharyngeal swab samples, we also recruited healthy children under 5 years of age from two of the 16 communes in the study catchment area in July 2008 using census data [18]. The inclusion criteria for the healthy children were a lack of fever, signs of respiratory infections or history of taking antimicrobials within 1 month prior to enrollment. The caregivers provided written informed consent. Information about potential risk factors was gathered using questionnaires (the questionnaire for data collection was submitted as Additional file 1: Figure S1).
Nasopharyngeal swab samples were collected following the standardized methods provided by the WHO working group [19]. Each normal saline (1 ml)-suspended sample was divided into two aliquots. One aliquot was used for bacterial culture and antimicrobial susceptibility testing. The second aliquot was processed using molecular techniques, including identification of pneumococcus by lytA or cpsA polymerase chain reaction (PCR) and molecular serotyping. A PCR assay targeting lytA was done as part of a multiplex PCR to detect three major respiratory pathogens (S. pneumoniae, Haemophilus influenaze and Moraxella catarrhalis). The PCR was performed in 25 μl volume; each reaction tube contained 5 × PCR buffer, 200 μM of deoxynucleoside triphosphate, 1.5 mM of MgCl2, 1.0 U of Go Taq DNA polymerase (Promega, Madison, WI) and 0 .5μM primers. 6 μl of crude extract was used as the DNA template. Thermal cycling was performed under the following condition: 94 °C for 5 min followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s. PCR assays targeting cpsA were done as part of 9 multiplex PCR assays for molecular serotyping. The first reaction contained primers for serotypes 14, 6, 19F, 23F, 11A, and the universal capsular primers (cpsA-F and cpsA-R). This was followed by the second reaction for serotypes 10F/10C/33C, 34, 15B/C, 19A, and 23A. 3 μl of DNA template was used. Thermal cycling was performed under the following condition: 94 °C for 3 min followed by 30 cycles of 94 °C for 45 s, 54 °C for 45 s, and 72 °C for 60 s. The primers used for the lytA and cpsA assays were lytA-F 5′-TCGTGCGTTTTAATTCCAGC, lytA-R 5′-TGAGGGACTACCGCCTTTAT, cpsA-F 5′-GCAGTACAGCAGTTTGTTGGACTGACC and cpsA-R 5′-GAATATTTTCATTATCAGTCCCAGTC. Details of the sample treatment were described elsewhere [20].
Susceptibility testing [21]
Nasopharyngeal swab samples were subjected to bacterial culture on site. A bacterial culture was performed on trypticase soy agar (Becton Dickinson and Company, Sparks, NV) with 7% defibrinated rabbit blood. Serial dilution (1:102 to 107) was performed before streaking to obtain single colonies. The plates were incubated in CO2 incubators at the microbiology laboratory at the study hospital. Potential S. pneumoniae colonies were selected based on colony morphology (alpha-hemolytic soft colonies with a central dimple [22]) and confirmed by optochin susceptibility. A bile solubility test was used if the optochin test was inconclusive.
Susceptibility testing was performed subsequently in a research laboratory. The minimum inhibitory concentrations (MICs) of 20 antimicrobial agents against S. pneumoniae isolates were determined using the agar dilution method with Mueller-Hinton agar (Becton Dickinson and Company, Sparks, NV) supplemented with 5% defibrinated horse blood. The antimicrobials used in the present study were penicillin (PEN) (Meiji Seika, Tokyo, Japan), amoxicillin (Sigma-Aldrich, St. Louis, MO), ampicillin, amoxicillin/clavulanate (Wako, Osaka, Japan), cefaclor (Shionogi, Osaka, Japan), cefuroxime (Shin Nihon Jitsugyo, Tokyo, Japan), cefotaxime (CTX) (Sigma-Aldrich, St. Louis, MO), cefepime (U.S. Pharmacopeia, Rockville, MD), imipenem, meropenem (MEM), erythromycin (Wako, Osaka, Japan), azithromycin (LKT Laboratories, St. Paul, MN), clarithromycin (Taisho Pharmaceutical, Tokyo, Japan), clindamycin, tetracycline, chloramphenicol (Wako, Osaka, Japan), trimethoprim/sulfamethoxazole (Sigma-Aldrich, St. Louis, MO), ofloxacin, rifampicin (Daiichi Pharmaceutical, Tokyo, Japan), and vancomycin (Shionogi, Osaka, Japan). For PEN and CTX, doubling dilutions over the concentration range of 0.004 to 128 μg/ml. For the other antimicrobials, appropriate breakpoint concentrations were used. The results were interpreted according to the breakpoints of the Clinical and Laboratory Standards Institute (CLSI) criteria (2016) [23]. S. pneumoniae ATCC® 49619 was used as the quality control strain.
Serotyping
A multiplex-PCR method that was described elsewhere [24, 25] was applied to confirm the pneumococcal serotypes. A non-typeable (NT) status was assigned for lytA-positive samples that were cpsA negative.
Genotyping of antimicrobial resistance genes
To identify PEN and other beta-lactam resistance genes in the pneumococcal culture-positive samples, we performed PCR with three primer sets (pbp1A, pbp2b and pbp2x) designed for PEN susceptible strains (Wakunaga Pharmaceutical, Osaka, Japan) according to the manufacturer’s instructions [26]. Macrolide-resistant genes (mefA and ermB) were also amplified [27].
Analysis
S. pneumoniae carriage was defined as a nasopharyngeal sample that was culture positive for S. pneumoniae and lytA positive, or culture positive for S. pneumoniae and cpsA positive. Logistic regression analyses were performed to identify risk factors for pneumococcal carriage and carriage of non-susceptible isolates.
Results
Pneumococcal carriage
In total, 883 nasopharyngeal samples were collected from 552 ARI cases and 331 healthy children. Potential S. pneumoniae isolates were obtained in 343 cases (223 ARI cases and 120 healthy children). Of those cases, lytA and cpsA positive results were observed in 266 (77.6%) and 277 (80.8%), respectively. In 31 cases (9.0%), lytA positive results were not obtained but cpsA genes were amplified; consequently, S. pneumoniae carriage was confirmed by the detection of the lytA or cpsA genes in 297 cases (33.6%, 95% confidence interval (CI) 30.6–36.8). Of those cases, 202 (36.6%, 32.7–40.7) were ARI cases, and 95 (28.7%, 24.1–33.8) were healthy children.
Table 1 shows the baseline characteristics of children with and without S. pneumoniae carriage and the risk factors for carriage. S. pneumoniae carriage varied significantly by age group; carriage was significantly higher in ARI cases than in healthy children in the 1–2 yr. and ≥ 3 yr. age groups but not among infants (6–11 m) (Additional file 2: Figure S2). In the multiple logistic regression analysis, the age group and daycare attendance were significantly associated with S. pneumoniae carriage. Although fluctuation of the carriage rate was observed throughout the year [28], the carriage rates were not significantly different between the four periods (Apr-Jun 2008, Jul-Sep 2008, Oct-Dec 2008 and Jan-Mar 2009) among children with ARIs in this study. The carriage rates were also not significantly different between 3 months (Apr 2008, Mar 2009 and Jul 2009). Consequently, we performed analyses using data obtained from healthy children in July 2008 and ARI cases throughout the entire year.
Antimicrobial susceptibility testing
The susceptibility patterns and the MIC50 and MIC90 values against the S. pneumoniae isolates from the study participants for 20 antimicrobials were interpreted according to the CLSI breakpoints (Table 2, Additional file 3: Table S1, and Additional file 4: Table S2). Alarmingly high MIC values for multiple beta-lactam antimicrobials, including carbapenems, were found among the S. pneumoniae isolates from both the ARI cases and healthy children. The percentages of S. pneumoniae isolates non-susceptible to PEN (non-meningitis breakpoint: ≥4 μg/ml), CTX (non-meningitis breakpoint: ≥2 μg/ml) and MEM (≥0.5 μg/ml) were 18.0, 25.8 and 75.6%, respectively. A high rate of non-susceptibility to the other antibiotic groups was also observed. S. pneumoniae isolates fully resistant to three or more antibiotic groups (multidrug-resistant) accounted for 94.9% of the total isolates if resistance to PEN was defined as an MIC ≥8 μg/ml.
To confirm our results of antimicrobial susceptibility using the agar dilution method, 43 samples were selected from our study, and the microdilution method was performed according to the CLSI guidelines [23, 29]. The percentages of S. pneumoniae isolates non-susceptible to PEN, CTX and MEM among the 43 isolates were 30.2% vs 32.6, 41.9% vs 62.8 and 86.0% vs 86.0% (agar dilution vs microdilution), respectively.
Risk of carrying isolates non-susceptible to multiple beta-lactams
A considerable number of the isolates had high MICs against multiple beta-lactam antimicrobials. An isolate that was non-susceptible to multiple beta-lactam antimicrobials was defined as an isolate with a PEN MIC ≥4 μg/ml, CTX MIC ≥2 μg/ml and MEM MIC ≥0.5 μg/ml; these isolates were further analyzed. These isolates accounted for 13.6% (95% CI: 10.1–17.9) of all isolates, and almost all of the isolates (38/40 isolates, 95.0%) were also non-susceptible to all of the macrolides (erythromycin, clarithromycin, and azithromycin), clindamycin, tetracycline and trimethoprim/sulfamethoxazole. We analyzed the risk or relevant factors for carriage of isolates non-susceptible to multiple beta-lactam antimicrobials (Table 3) and found that daycare attendance was the only significant risk factor. None of the other factors, including ARI hospitalization, age group, or prior antimicrobial use, were significantly associated.
Serotyping
Serotyping results were available for 92 healthy children and 202 ARI cases. Serogroup 6 was most common in both groups, followed by 19F (Additional file 5: Figure S3). The PCV-13 serotypes accounted for 80.4 and 72.3% of the isolates from the healthy children and ARI cases, respectively.
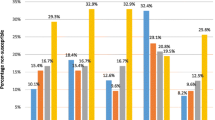
The proportions of S. pneumoniae isolates non-susceptible to PEN, CTX, MEM and all these three beta-lactam antimicrobials by serotypes/serogroups are shown in Fig. 1a-d and Additional file 6: Table S3. Serotype 19F was significantly associated with the carriage of S. pneumoniae non-susceptible to PEN (p < 0.0001), CTX (p = 0.0002), MEM (p = 0.0005) and all three beta-lactam antimicrobials (p < 0.0001), and serogroup 6 was significantly associated with the carriage of S. pneumoniae non-susceptible to CTX (p = 0.0468) and MEM (p = 0.0187). The detection of pneumococcal isolates non-susceptible to MEM was also significantly associated with serotypes 23F (p = 0.0029) and 11A (p = 0.0397). Of the S. pneumoniae isolates non-susceptible to all three beta-lactam antimicrobials, the majority (90.0%) were of the PCV13-vaccine serotype.
The proportions of S. pneumoniae isolates non-susceptible to beta-lactam antimicrobials by serotypes/serogroups (a: PEN, b: CTX, c: MEM, d: multiple beta-lactam antimicrobials). Non-susceptible isolates (PEN MIC ≥4 μg/ml, CTX MIC ≥2 μg/ml, MEM MIC ≥0.5 μg/ml and multiple beta-lactams) are shown in red. *: includes serotypes 23A, 29, and 34. The analysis was performed using pneumococcal isolates with a single serotype by multiplex-PCR (n = 285). #1: p < 0.0001, #2: p = 0.0002, #3: p = 0.0468, #4: p = 0.0005, #5: p = 0.0187, #6: p = 0.0029, #7: p = 0.0397
Genotyping of antimicrobial resistance genes
The majority of the S. pneumoniae-positive samples (265/290, 91.4%: 95% CI 87.6–94.1) were also positive for genotypic penicillin-resistant S. pneumoniae (gPRSP) (pbp1a + 2x + 2b). The genotypic penicillin-sensitive S. pneumoniae (gPSSP) samples accounted for only 2.4% (95% CI: 1.17–4.90) of the isolates. A total of 282 samples (97.2%: 95% CI 94.7–98.6) were positive for either ermB or mefA. Details are shown in Additional file 7: Table S4 and Additional file 8: Table S5.
The impact of pneumococci non-susceptible to multiple beta-lactam antimicrobials on clinical indices
Clinical indices were evaluated for ARI cases where pneumococci were detected (n = 202). The detection of pneumococci non-susceptible to multiple beta-lactam antimicrobials (n = 18) among the ARI cases did not lead to significant prolongation of hospitalization and did not affect the type of antimicrobials administered during the hospital stay.
Discussion
This study is the first to report that an alarmingly high proportion of children in Vietnam carry S. pneumoniae isolates non-susceptible to multiple beta-lactam antimicrobials, including carbapenems. These high levels of non-susceptibility were observed regardless of whether the child was sick with an ARI in the hospital or healthy in the community. These results were obtained using a population-based study at a site in central Vietnam. The proportions of non-susceptible pneumococcal isolates with high MICs (cefotaxime or ceftriaxone MIC ≥2 μg/ml) were 0–4.4% in previous studies conducted in Vietnam, even among clinical isolates [6,7,8,9,10]. Although a few studies demonstrated significant increases in pneumococci non-susceptible to cefotaxime among nasopharyngeal carriage isolates, the target populations of these studies were daycare attendees or children who visited a clinic (including health check visits) [30,31,32], who were not necessarily representative of healthy children living in the community. The present study clearly showed that S. pneumoniae strains non-susceptible to multiple beta-lactam antimicrobials were circulating deep in the community of central Vietnam.
The proportion of pneumococcal isolates non-susceptible to multiple beta-lactam antimicrobials was not affected by prior antimicrobial use. The association between resistant pneumococci (both colonization and clinical samples) and the area of consumption of antimicrobials has been described in an ecological study [33], a cross-sectional study [34] and a cluster-randomized trial [35], suggesting the importance of antibiotic selection pressure in the community. The relationship between resistance and individual consumption of antimicrobials has also been reported in a cross-sectional study [34], a non-randomized observational study [36], and a case-controlled study [37]. In these study designs, bias is a matter of concern. The impact of macrolide therapy on pharyngeal carriage of macrolide-resistant oral streptococci was investigated in a randomized, double-blind, placebo-controlled study in which healthy volunteers were treated with azithromycin and clarithromycin [38], but no such studies have been performed for S. pneumoniae. Once resistant clones become predominant in a community and account for most of the circulating isolates, healthy children most likely acquire resistant isolates even without taking antimicrobials. This process may occur in daycare centers, as shown in the current study. Indeed, Bartoloni et al. reported the dissemination of antimicrobial-resistant commensal Escherichia coli without heavy exposure to antimicrobials in a remote Bolivian area [39].
Our data demonstrated that a specific serotype (19F) was primarily responsible for non-susceptibility. The disproportionate distribution of serotypes among non-susceptible isolates was consistent with previous observations of the emergence of globally distributed resistant clones [40,41,42]. Spain23F-1, Taiwan19F-14 and their related strains have been described in Vietnam [7, 9]. Further molecular analysis using multi-locus sequence typing (MLST) [43] will demonstrate the presence of globally circulating resistant clones. Molecular analysis will also be a clue to the mystery: why were S. pneumoniae strains non-susceptible to carbapenems circulating in Vietnam without the overuse of carbapenems. In a small-scale hospital-based study conducted at the study site in 2011, third generation cephalosporins were frequently used for pneumonia, whereas oral second-generation cephalosporins and penicillins were prescribed for milder cases; carbapenems were not selected at all (Toizumi M, et al. unpublished data).
One solution for the high rate of antimicrobial resistance is the introduction of vaccines. The proportion of PCV-13 serotypes among the carriage isolates was greater than 70% for both the healthy children and the ARI cases, similar to reports from developed countries in the pre-PCV era [14, 44]. Furthermore, PCV13 vaccine serotypes accounted for 90% of the pneumococcal isolates non-susceptible to multiple beta-lactam antimicrobials. Introduction of a vaccine is expected to increase the susceptibility of the circulating strains. Genotypic analysis revealed that gPSSP expression accounted for less than 3% of the pneumococcal isolates and that 97.6% of the isolates possessed at least one resistant pbp gene. Relatively drug-susceptible pneumococci with non-vaccine serotypes may exchange resistance genes through genetic recombination and acquire higher levels of resistance unless appropriate regulation of antimicrobials (reduction of the area of consumption of antimicrobials) is implemented. In a surveillance study of healthcare utilization conducted in the study area in 2006, approximately 40% of 1355 caregivers of ARI children visited a pharmacy for their first choice of healthcare provider and slightly less than 60% of them purchased antimicrobials (Toizumi M, et al. unpublished data). PCV-13 is expected to reduce the non-susceptible S. pneumoniae carriage to a greater extent than PCV-7 [15]. However, careful monitoring of emerging mutants (serotype switching or acquisition of drug resistance genes) is required [45, 46].
We did not detect a clinical impact of the carriage of pneumococcal isolates non-susceptible to multiple beta-lactam antimicrobials. The detection of these non-susceptible pneumococcal isolates in the ARI cases did not lead to significant prolongation of hospitalization. One possible reason was that our study population did not include severe cases who were admitted to the pediatric intensive care unit. Feikin et al. described the positive association between mortality from invasive pneumococcal pneumonia and PEN MICs ≥4 μg/ml (Odds Ratio: 7.1) or cefotaxime MICs ≥2 μg/ml (Odds Ratio: 5.9) after the first 4 hospital days [47].
The current study has some limitations. Information on sibling health conditions was not obtained from the questionnaires; therefore, whether the siblings of the healthy children took antimicrobials during the study period and whether intra-familial transmission occurred were unknown. A previous study in isolated, rural Utah communities demonstrated an association between the detection of non-susceptible S. pneumoniae and having a sibling colonized with resistant pneumococci as evidence of intra-familial transmission based on pulsed field gel electrophoresis [48].
Our method of pneumococcal isolation (based upon colony morphology on a non-selective medium) and identification (optochin susceptibility and a bile solubility test) could underestimate NT pneumococcal carriage status because of their morphological differences (less conspicuous dimples of the colonies) from serotypeable S. pneumoniae. We considered only bile soluble isolates as pneumococci, which may also lead to the underestimation of NT pneumococcal carriage. Some other members of the viridans group of streptococci might be optochin susceptible which lead to false positive results; to avoid the problem, we defined S. pneumoniae carriage as a nasopharyngeal sample was culture positive for S. pneumoniae and lytA positive, or culture positive for S. pneumoniae and cpsA positive.
The agar dilution method used for MIC testing may not be the standard method used in many laboratories. However, we found good concordance between the agar dilution method and the microdilution method. Therefore, we believe that the significant increase in non-susceptibility to PEN and other beta-lactam antimicrobials in the present study is a reliable finding.
In Vietnam, the PCV has not been introduced yet and irrational use of antimicrobials is a great issue. Thus data from this study will give us background information to compare with the ongoing study on current situation of antibiotic resistance pattern, serotype predominance, and sequence type distribution of pneumococci in Vietnam.
Conclusion
S. pneumoniae serotype 19F isolates non-susceptible to multiple beta-lactam antimicrobials are widely circulating among children living in central Vietnam. The introduction of a PCV is expected to improve antimicrobial susceptibility.
Abbreviations
- ARI:
-
Acute respiratory infection
- CLSI:
-
Clinical and Laboratory Standards Institute
- CTX:
-
Cefotaxime
- gPRSP:
-
genotypic penicillin-resistant S. pneumoniae
- gPSSP:
-
genotypic penicillin-sensitive S. pneumoniae
- IPD:
-
Invasive pneumococcal disease
- MEM:
-
Meropenem
- MIC:
-
Minimum inhibitory concentration
- MLST:
-
multi-locus sequence typing
- PCR:
-
Polymerase chain reaction
- PCV:
-
Pneumococcal conjugate vaccine
- PEN:
-
Penicillin
- PRSP:
-
penicillin-resistant Streptococcus pneumoniae
References
Walker CL, Rudan I, Liu L, Nair H, Theodoratou E, Bhutta ZA, O’Brien KL, Campbell H, Black RE. Global burden of childhood pneumonia and diarrhoea. Lancet. 2013;381:1405–16. https://doi.org/10.1016/S0140-6736(13)60222-6.
Hicks LA, Harrison LH, Flannery B, Hadler JL, Schaffner W, Craig AS, Jackson D, Thomas A, Beall B, Lynfield R, Reingold A, Farley MM, Whitney CG, for the Active Bacterial Core Surveillance Program of the Emerging Infections Program Network. Incidence of pneumococcal disease due to non-pneumococcal conjugate vaccine (PCV7) serotypes in the United States during the era of widespread PCV7 vaccination, 1998-2004. J Infect Dis. 2007;196:1346–54. https://doi.org/10.1086/521626.
Whitney CG, Farley MM, Hadler J, Harrison LH, Bennett NM, Lynfield R, Reingold A, Cieslak PR, Pilishvili T, Jackson D, Facklam RR, Jorgensen JH, Schuchat A, Active Bacterial Core Surveillance of the Emerging Infections Program Network. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N Engl J Med. 2003;348:1737–46. https://doi.org/10.1056/NEJMoa022823.
Simonsen L, Taylor RJ, Young-Xu Y, Haber M, May L, Klugman KP. Impact of pneumococcal conjugate vaccination of infants on pneumonia and influenza hospitalization and mortality in all age groups in the United States. mBio. 2011;2:e00309–10. https://doi.org/10.1128/mBio.00309-10.
Grijalva CG, Nuorti JP, Arbogast PG, Martin SW, Edwards KM, Griffin MR. Decline in pneumonia admissions after routine childhood immunisation with pneumococcal conjugate vaccine in the USA: a time-series analysis. Lancet. 2007;369:1179–86. https://doi.org/10.1016/S0140-6736(07)60564-9.
Parry CM, Diep TS, Wain J, Hoa NTT, Gainsborough M, Nga D, Davies C, Phu NH, Hien TT, White NJ, Farrar JJ. Nasal carriage in Vietnamese children of Streptococcus pneumoniae resistant to multiple antimicrobial agents. Antimicrob Agents Chemother. 2000;44:484–8. https://doi.org/10.1128/AAC.44.3.484-488.2000.
Schultsz C, Vien le M, Campbell JI, Chau NVV, Diep TS, Hoang NVM, Nga TTT, Savelkoul P, Stepnieuwska K, Parry C, Hien TT, Farrar JJ. Changes in the nasal carriage of drug-resistant Streptococcus pneumoniae in urban and rural Vietnamese schoolchildren. Trans R Soc Trop Med Hyg. 2007;101:484–92. https://doi.org/10.1016/j.trstmh.2006.08.010.
Hoa NQ, Trung NV, Larsson M, Eriksson B, Phuc HD, Chuc NT, Lundborg CS. Decreased Streptococcus pneumoniae susceptibility to oral antibiotics among children in rural Vietnam: a community study. BMC Infect Dis. 2010;10:85. https://doi.org/10.1186/1471-2334-10-85.
Parry CM, Duong NM, Zhou J, Mai NTH, Diep TS, Thinh LQ, Wain J, Chau NVV, Griffiths D, Day NPJ, White NJ, Hien TT, Spratt BG, Farrar J. Emergence in Vietnam of Streptococcus pneumoniae resistant to multiple antimicrobial agents as a result of dissemination of the multiresistant Spain23F-1 clone. Antimicrob Agents Chemother. 2002;46:3512–7.
Kim SH, Song JH, Chung DR, Thamlikitkul V, Yang Y, Wang H, Lu M, So TM, Hsueh PR, Yasin RM, Carlos CC, Pham HV, Lalitha MK, Shimono N, Perera J, Shibl AM, Baek JY, Kang CI, Ko KS, Peck KR, ANSORP Study Group. Changing trends in antimicrobial resistance and serotypes of Streptococcus pneumoniae isolates in Asian countries: an Asian network for surveillance of resistant pathogens (ANSORP) study. Antimicrob Agents Chemother. 2012;56:1418–26. https://doi.org/10.1128/AAC.05658-11.
Kyaw MH, Lynfield R, Schaffner W, Craig AS, Hadler J, Reingold A, Thomas AR, Harrison LH, Bennett NM, Farley MM, Facklam RR, Jorgensen JH, Besser J, Zell ER, Schuchat A, Whitney CG, Active Bacterial Core Surveillance of the Emerging Infections Program Network. Effect of introduction of the pneumococcal conjugate vaccine on drug-resistant Streptococcus pneumoniae. N Engl J Med. 2006;354:1455–63. https://doi.org/10.1056/NEJMoa051642.
Huang SS, Platt R, Rifas-Shiman SL, Pelton SI, Goldmann D, Finkelstein JA. Post-PCV7 changes in colonizing pneumococcal serotypes in 16 Massachusetts communities, 2001 and 2004. Pediatrics. 2005;116:e408–13. https://doi.org/10.1542/peds.2004-2338.
Moore MR, Gertz RE Jr, Woodbury RL, Barkocy-Gallagher GA, Schaffner W, Lexau C, Gershman K, Reingold A, Farley M, Harrison LH, Hadler JL, Bennett NM, Thomas AR, McGee L, Pilishvili T, Brueggemann AB, Whitney CG, Jorgensen JH, Beall B. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J Infect Dis. 2008;197:1016–27. https://doi.org/10.1086/528996.
Brueggemann AB, Griffiths DT, Meats E, Peto T, Crook DW, Spratt BG. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J Infect Dis. 2003;187:1424–32. https://doi.org/10.1086/374624.
Dagan R, Juergens C, Trammel J, Patterson S, Greenberg D, Givon-Lavi N, Porat N, Gurtman A, Gruber WC, Scott DA. Efficacy of 13-valent pneumococcal conjugate vaccine (PCV13) versus that of 7-valent PCV (PCV7) against nasopharyngeal colonization of antibiotic-nonsusceptible Streptococcus pneumoniae. J Infect Dis. 2015;211:1144–53. https://doi.org/10.1093/infdis/jiu576.
Yoshida LM, Suzuki M, Thiem VD, Smith WP, Tsuzuki A, Huong VTT, Takahashi K, Miyakawa M, Anh NTH, Watanabe K, Ai NTT, Tho le H, Kilgore P, Yoshino H, Toizumi M, Yasunami M, Moriuchi H, Anh DD, Ariyoshi K. Population based cohort study for pediatric infectious diseases research in Vietnam. Trop Med Health. 2014;42:47–58. https://doi.org/10.2149/tmh.2014-S07.
Yoshida LM, Suzuki M, Yamamoto T, Nguyen HA, Nguyen CD, Nguyen AT, Oishi K, Vu TD, Le TH, Le MQ, Yanai H, Kilgore PE, Dang DA, Ariyoshi K. Viral pathogens associated with acute respiratory infections in central Vietnamese children. Pediatr Infect Dis J. 2010;29:75–7. https://doi.org/10.1097/INF.0b013e3181af61e9.
Suzuki M, Thiem VD, Yanai H, Matsubayashi T, Yoshida LM, Tho LH, Minh TT, Anh DD, Kilgore PE, Ariyoshi K. Association of environmental tobacco smoking exposure with an increased risk of hospital admissions for pneumonia in children under 5 years of age in Vietnam. Thorax. 2009;64:484–9. https://doi.org/10.1136/thx.2008.106385.
O’brien KL, Nohynek H, World Health Organization Pneumococcal Vaccine Trials Carriage Working Group. Report from a WHO working group: standard method for detecting upper respiratory carriage of Streptococcus pneumoniae. Pediatr Infect Dis J. 2003;22:e1–11. https://doi.org/10.1097/01.inf.0000049347.42983.77.
Dhoubhadel BG, Yasunami M, Nguyen HAT, Suzuki M, Vu TH, Thi Thuy Nguyen ATT, Dang DA, Yoshida LM, Ariyoshi K. Bacterial load of pneumococcal serotypes correlates with their prevalence and multiple serotypes is associated with acute respiratory infections among children less than 5 years of age. PLoS One. 2014;9:e110777. https://doi.org/10.1371/journal.pone.0110777.
Watanabe K, Anh DD, Huong Ple T, Nguyet NT, Anh NTH, Thi NT, Dung NT, Phuong DM, Rusizoka OS, Nagatake T, Watanabe H, Oishi K. Drug-resistant pneumococci in children with acute lower respiratory infections in Vietnam. Pediatr Int. 2008;50:514–8. https://doi.org/10.1111/j.1442-200X.2008.02616.x.
Werno AM, Murdoch DR. Laboratory diagnosis of invasive pneumococcal disease. Clin Infect Dis. 2008;46:926–32. https://doi.org/10.1086/528798.
Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. Wayne: CLSI; 2016.
Vu HTT, Yoshida LM, Suzuki M, Nguyen HAT, Nguyen CDL, Nguyen ATT, Oishi K, Yamamoto T, Watanabe K, Vu TD. Association between nasopharyngeal load of Streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatr Infect Dis J. 2011;30:11–8. https://doi.org/10.1097/INF.0b013e3181f111a2.
Pai R, Gertz RE, Beall B. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol. 2006;44:124–31. https://doi.org/10.1128/JCM.44.1.124-131.200642.6.2518-2522.2004.
Ubukata K, Chiba N, Hasegawa K, Kobayashi R, Iwata S, Sunakawa K. Antibiotic susceptibility in relation to penicillin-binding protein genes and serotype distribution of Streptococcus pneumoniae strains responsible for meningitis in Japan, 1999 to 2002. Antimicrob Agents Chemother. 2004;48:1488–94. https://doi.org/10.1128/AAC.48.5.1488-1494.2004.
Ubukata K, Iwata S, Sunakawa K. In vitro activities of new ketolide, telithromycin, and eight other macrolide antibiotics against Streptococcus pneumoniae having mefA and ermB genes that mediate macrolide resistance. J Infect Chemother. 2003;9:221–6. https://doi.org/10.1007/s10156-003-0258-2.
Syrjänen RK, Kilpi TM, Kaijalainen TH, Herva EE, Takala AK. Nasopharyngeal carriage of Streptococcus pneumoniae in Finnish children younger than 2 years old. J Infect Dis. 2001;184:451–9. https://doi.org/10.1086/322048.
Clinical and Laboratory Standards Institute (CLSI). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standards tenth edition. Wayne: CLSI; 2015.
Chiu SS, Ho PL, Chow FKH, Yuen KY, Lau YL. Nasopharyngeal carriage of antimicrobial-resistant Streptococcus pneumoniae among young children attending 79 kindergartens and day care centers in Hong Kong. Antimicrob Agents Chemother. 2001;45:2765–70. https://doi.org/10.1128/AAC.45.10.2765-2770.2001.
Kuo CY, Hwang KP, Hsieh YC, Cheng CH, Huang FL, Shen YH, Huang YC, Chiu CH, Chen PY, Lin TY. Nasopharyngeal carriage of Streptococcus pneumoniae in Taiwan before and after the introduction of a conjugate vaccine. Vaccine. 2011;29:5171–7. https://doi.org/10.1016/j.vaccine.2011.05.034.
Finkelstein JA, Huang SS, Daniel J, Rifas-Shiman SL, Kleinman K, Goldmann D, Pelton SI, DeMaria A, Platt R. Antibiotic-resistant Streptococcus pneumoniae in the heptavalent pneumococcal conjugate vaccine era: predictors of carriage in a multicommunity sample. Pediatrics. 2003;112:862–9. https://doi.org/10.1542/peds.112.4.862.
SLAM B, Cars O, Buchholz U, Mölstad S, Goettsch W, Veldhuijzen IK, Kool JL, MJW S, Degener JE, European Antimicrobial Resistance Surveillance System. A European study on the relationship between antimicrobial use and antimicrobial resistance. Emerg Infect Dis. 2002;8:278–82. https://doi.org/10.3201/eid0803.010192.
Arason VA, Kristinsson KG, Sigurdsson JA, Stefánsdóttir G, Mölstad S, Gudmundsson S. Do antimicrobials increase the carriage rate of penicillin resistant pneumococci in children? Cross sectional prevalence study. BMJ. 1996;313:387–91. https://doi.org/10.1136/bmj.313.7054.387.
Skalet AH, Cevallos V, Ayele B, Gebre T, Zhou Z, Jorgensen JH, Zerihun M, Habte D, Assefa Y, Emerson PM, Gaynor BD, Porco TC, Lietman TM, Keenan JD. Antibiotic selection pressure and macrolide resistance in nasopharyngeal Streptococcus pneumoniae: a cluster-randomized clinical trial. PLoS Med. 2010;7:e1000377. https://doi.org/10.1371/journal.pmed.1000377.
Feikin DR, Dowell SF, Nwanyanwu OC, Klugman KP, Kazembe PN, Barat LM, Graf C, Bloland PB, Ziba C, Huebner RE, Schwartz B. Increased carriage of trimethoprim/sulfamethoxazole-resistant Streptococcus pneumoniae in Malawian children after treatment for malaria with sulfadoxine/pyrimethamine. J Infect Dis. 2000;181:1501–5. https://doi.org/10.1086/315382.
Metlay JP, Fishman NO, Joffe MM, Kallan MJ, Chittams JL, Edelstein PH. Macrolide resistance in adults with bacteremic pneumococcal pneumonia. Emerg Infect Dis. 2006;12:1223–30. https://doi.org/10.3201/eid1208.060017.
Malhotra-Kumar S, Lammens C, Coenen S, Van Herck K, Goossens H. Effect of azithromycin and clarithromycin therapy on pharyngeal carriage of macrolide-resistant streptococci in healthy volunteers: a randomised, double-blind, placebo-controlled study. Lancet. 2007;369:482–90. https://doi.org/10.1016/S0140-6736(07)60235-9.
Bartoloni A, Bartalesi F, Mantella A, Dell’Amico E, Roselli M, Strohmeyer M, Barahona HG, Barrón VP, Paradisi F, Rossolini GM. High prevalence of acquired antimicrobial resistance unrelated to heavy antimicrobial consumption. J Infect Dis. 2004;189:1291–4. https://doi.org/10.1086/382191.
Richter SS, Heilmann KP, Dohrn CL, Riahi F, Beekmann SE, Doern GV. Changing epidemiology of antimicrobial-resistant Streptococcus pneumoniae in the United States, 2004-2005. Clin Infect Dis. 2009;48:e23–ee33. https://doi.org/10.1086/595857.
Sharma D, Baughman W, Holst A, Thomas S, Jackson D, da Gloria Carvalho MG, Beall B, Satola S, Jerris R, Jain S, Farley MM, Nuorti JP. Pneumococcal carriage and invasive disease in children before introduction of the 13-valent conjugate vaccine: comparison with the era before 7-valent conjugate vaccine. Pediatr Infect Dis J. 2013;32:e45–ee53. https://doi.org/10.1097/INF.0b013e3182788fdd.
McGee L, McDougal L, Zhou J, Spratt BG, Tenover FC, George R, Hakenbeck R, Hryniewicz W, Lefévre JC, Tomasz A, Klugman KP. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J Clin Microbiol. 2001;39:2565–71. https://doi.org/10.1128/JCM.39.7.2565-2571.2001.
Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–60. https://doi.org/10.1099/00221287-144-11-3049.
Bogaert D, de Groot R, Hermans PWM. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis. 2004;4:144–54. https://doi.org/10.1016/S1473-3099(04)00938-7.
Pillai DR, Shahinas D, Buzina A, Pollock RA, Lau R, Khairnar K, Wong A, Farrell DJ, Green K, McGeer A, Low DE. Genome-wide dissection of globally emergent multi-drug resistant serotype 19A Streptococcus pneumoniae. BMC Genomics. 2009;10:642. https://doi.org/10.1186/1471-2164-10-642.
Croucher NJ, Chewapreecha C, Hanage WP, Harris SR, McGee L, van der Linden M, Song JH, Ko KS, de Lencastre H, Turner C, Yang F, Sá-Leão R, Beall B, Klugman KP, Parkhill J, Turner P, Bentley SD. Evidence for soft selective sweeps in the evolution of pneumococcal multidrug resistance and vaccine escape. Genome Biol Evol. 2014;6:1589–602. https://doi.org/10.1093/gbe/evu120.
Feikin DR, Schuchat A, Kolczak M, Barrett NL, Harrison LH, Lefkowitz L, McGeer A, Farley MM, Vugia DJ, Lexau C, Stefonek KR, Patterson JE, Jorgensen JH. Mortality from invasive pneumococcal pneumonia in the era of antibiotic resistance, 1995-1997. Am J Public Health. 2000;90:223–9. https://doi.org/10.2105/AJPH.90.2.223.
Samore MH, Magill MK, Alder SC, Severina E, Morrison-de Boer L, Lyon JL, Carroll K, Leary J, Stone MB, Bradford D, Reading J, Tomasz A, Sande MA. High rates of multiple antibiotic resistance in Streptococcus pneumoniae from healthy children living in isolated rural communities: association with cephalosporin use and intrafamilial transmission. Pediatrics. 2001;108:856–65. https://doi.org/10.1542/peds.108.4.856.
Acknowledgments
We thank Ai Thi Thuy Nguyen, the staff members of Khanh Hoa General Hospital, Tho Huu Le and staff of Khanh Hoa Health Service for their help with the study. We also thank Kiwao Watanabe, Kyoko Uchibori and Stephanie Jane Airs for technical assistance with susceptibility testing. We are grateful to Michiko Toizumi for providing us useful data on antimicrobial consumption in the study area.
Funding
This research is supported by the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) from Ministry of Education, Culture, Sport, Science & Technology in Japan, and Japan Agency for Medical Research and Development (AMED). The funding body had no role in the design of the study and collection, analysis, and interpretation of data, and in writing the manuscript.
Availability of data and materials
The datasets generated and analyzed during the current study is presented in Additional file 9.
Author information
Authors and Affiliations
Contributions
LMY, ADD, and KA designed the study, coordinated the field work and supervised in writing the manuscript. HATN and HTTV did the data and sample collection, laboratory testing, data analysis and wrote the manuscript. HF and CMP did the statistical analysis. HATN drafted the manuscript with HF, CMP, KA, and LMY. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The research protocol was approved by the Nagasaki University Ethical Review Board, the National Institute of Hygiene and Epidemiology scientific review committee in Hanoi, and the Khanh Hoa Provincial Health Service ethical review board. Written informed consent was obtained from the parents or guardians prior to sample collection and the interviews.
Consent for publication
Not applicable.
Competing interests
The authors have no conflicts of interest to declare. The corresponding author LMY is an associate editor of BMC infectious Diseases Journal.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S1. The epidemiologic-clinical-laboratory form used in the study. (DOCX 276 kb)
Additional file 2:
Figure S2. Pneumococcal carriage rate by age group. (DOCX 31 kb)
Additional file 3:
Table S1. MIC Distribution against Pneumococcal Isolates from Study Participants. (DOCX 16 kb)
Additional file 4:
Table S2. Non-Susceptibility of Penicillin-Susceptible, -Intermediate, and -Resistant S. pneumoniae to Other Antimicrobials. (DOCX 16 kb)
Additional file 5:
Figure S3. Serotype distribution among pneumococcal carriage isolates from healthy children and ARI cases. (DOCX 26 kb)
Additional file 6:
Table S3. Multiple Beta-Lactam Non-Susceptible Isolates by Serotype/Serogroup. (DOCX 16 kb)
Additional file 7:
Table S4. pbp Genotypes. (DOCX 15 kb)
Additional file 8:
Table S5. Macrolide Resistance Genes. (DOCX 15 kb)
Additional file 9:
Data set used for analysis. (XLSX 98 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Nguyen, H.A.T., Fujii, H., Vu, H.T.T. et al. An alarmingly high nasal carriage rate of Streptococcus pneumoniae serotype 19F non-susceptible to multiple beta-lactam antimicrobials among Vietnamese children. BMC Infect Dis 19, 241 (2019). https://doi.org/10.1186/s12879-019-3861-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-019-3861-2