Abstract
Background
Infection with human immunodeficiency virus (HIV) influences the outcome and natural disease progression of hepatitis C virus (HCV) infection. While the majority of HCV mono-infected and HCV/HIV co-infected subjects develop chronic HCV infection, 20–46% of mono- and co-infected subjects spontaneously clear HCV infection. The mechanism underlying viral clearance is not clearly understood. Analysis of differential cellular gene expression (mRNA) between HIV-infected patients with persistent HCV infection or spontaneous clearance could provide a unique opportunity to decipher the mechanism of HCV clearance.
Methods
Plasma RNA from HIV/HCV co-infected subjects who cleared HCV and those who remained chronically infected with HCV was sequenced using Ion Torrent technology. The sequencing results were analyzed to identify transcripts that are associated with HCV clearance by measuring differential gene expression in HIV/HCV co-infected subjects who cleared HCV and those who remained chronically infected with HCV.
Results
We have identified plasma mRNA, the levels of which are significantly elevated (at least 5 fold, False Discovery Rate (FDR) <0.05) before HCV infection in subjects who cleared HCV compared to those who remained chronically infected. Upon further analysis of these differentially expressed genes, before and after HCV infection, we found that before HCV infection 12 genes were uniquely upregulated in the clearance group compared to the chronically infected group. Importantly, a number of these 12 genes and their upstream regulators (such as CCL3, IL17D, LBP, SOCS3, NFKBIL1, IRF) are associated with innate immune response functions.
Conclusions
These results suggest that subjects who spontaneously clear HCV may express these unique genes associated with innate immune functions.
Similar content being viewed by others
Background
Hepatitis C virus (HCV) infection is the leading cause of hepatocellular carcinoma in the United States [1]. Due to similar transmission routes, approximately one third of HIV-infected individuals are co-infected with HCV [2–4]. In HCV mono infection, nearly 70–80% of infected individuals develop chronic infection and liver cirrhosis, some of whom develop hepatocellular carcinoma, while 20–30% of infected individuals spontaneously clear the viral infection within one year of infection [5]. However, in HIV/HCV co-infected individuals, infection with HIV is associated with a decrease in the rate of spontaneous HCV clearance, exacerbates the natural course of HCV infection, and accelerates the development of HCC [6–10]. Currently, the mechanism of spontaneous HCV clearance and how HIV might affect this are not clear. Certain immunological components have been shown to be involved in HCV clearance [11–13]. An HCV-specific T-cell response in the liver has also been reported to be associated with HCV clearance. Various factors, such as gene polymorphisms in interleukin 28B (IL28B) and IP-10 have been implicated in mediating host susceptibility/resistance to HCV infection and disease progression [14–17]. In HCV-infected chimpanzees, interleukin binding factor 3 (ILF3) and cytotoxic granule associated cRNA binding protein (TIA1), which are associated with robust T-cell responses, were highly induced in animals who cleared the virus [18]. However, these represent only a few of the many factors that are potentially involved in HCV clearance.
Differential cellular gene expression in HIV-infected subjects has been shown to be associated with HIV disease progression [19]. Therefore, analysis of differential cellular gene expression between HIV infected patients with persistent HCV infection and those with spontaneous clearance of this virus could provide a unique opportunity to decipher the mechanism of HCV clearance in HIV/HCV co-infected subjects.
Pathogenic changes occurring in any tissues or organs are expected to leave footprints in the blood. Consequently, measurement of certain target RNA in the plasma has been explored or used for diagnosis of a number of diseases and cancers [20–24]. We hypothesize that the cellular (immunological and non-immunological) factors that are responsible for spontaneous HCV clearance leave footprints in the plasma, leading to differential plasma RNA expression profiles between those who spontaneously clear HCV infection and chronically infected individuals, with or without HIV co-infection. It is presumed that differential gene expression in plasma between those who cleared HCV and those who remained chronically infected will provide clues to the mechanism of HCV clearance. Due to the low quantity of RNA in blood plasma, currently there is no convenient way to characterize plasma RNA profiles. Recent advancement of next-generation sequencing technologies has made it possible for unbiased and comprehensive analysis of the gene expression from both cells and tissues. We have recently modified the NGS technology in Ion Proton platform and characterized RNA profiles in plasma.
In this study we identified plasma mRNA, the levels of which are significantly increased (at least 5 fold, FDR <0.05) in the HCV clearance group compared to chronically HCV infected patients. Upon further analysis of these differentially expressed genes, we have identified 12 genes that are upregulated only in clearance group before HCV infection. Moreover, some of these 12 genes and their upstream regulators are associated with innate immune functions.
Methods
Patients and samples
Frozen plasma samples were obtained from 13 HCV seroconverters who were infected with HIV for more than 10 years before HCV seroconversion in the Multicenter AIDS Cohort Study (MACS) [25]. MACS is the first and largest study specifically created to examine the natural history of AIDS. MACS participants are seen every six months and at each visit, plasma is taken for storage at -80 °C. HCV seroconversion is defined by the HCV antibody switching from negative to positive in plasma and the plasma HCV antibody positivity were persistent for all subsequent visits of the individuals. Furthermore, HCV seroconversion visits were confirmed by reverse transcriptase polymerase chain reaction specific for detecting HCV RNA. Spontaneous HCV clearance is defined as plasma HCV RNA were never detected or detected only one to two times around the time of seroconversion. Chronic HCV infection was defined as plasma HCV RNA being persistently detected for more than five-ten years. From five subjects with spontaneous HCV clearance and eight subjects with chronic HCV infection, we examined their plasma samples that were obtained immediately before and after HCV infection (MACS visits are six months apart). All HCV infections were reported to have been acquired by sexual transmission. At the time of HCV infection, all subjects were naïve to antiretroviral therapy and anti-HCV treatment.
Nucleic acid extraction, cDNA library construction and sequencing
Total nucleic acids were extracted from l ml of frozen cryopreserved plasma using an automated NucliSens EasyMag nucleic acid extraction machine (bioMérieux, Durham, NC) followed by removal of DNA from the nucleic extract with Qiagen AllPrep DNA/RNA mini kit and ribosomal RNA by a Low Input RiboMinus System (Life Technologies). The cDNA Library was then constructed using an Ion Torrent Total RNA-Seq Kit (Life Technologies) for whole transcriptome libraries and Barcodes 1 through 8 from an Ion Xpress 1–16 barcoding kit were used (Life Technologies) for each individual sample. cDNA libraries were quantified by qPCR using an Ion Library Quantitation Kit (Life Technologies) to determine a suitable template dilution factor for subsequent emulsion PCR and sequencing.
Four barcoded samples were combined for one sequencing reaction. Template preparation for sequencing was conducted using the OneTouch Ion™ Template Kit in the OneTouch machine (Life Technologies). Ion Torrent sequencing was conducted using the Ion Proton Sequencing Kit (Life Technologies) on an Ion Proton Machine (Life Technologies) using a P1(v2)-chip (Life Technologies). The constructed cDNA was also used for HIV viral load measurement using a quantitative-real time PCR for HIV gag RNA, with a sensitivity of 10 copies/mL [26].
Sequence analysis
Raw sequencing reads in FastQ format were assessed for quality using CLC Genomics Workbench 7. Reads were accepted based on the length (longer than 25 nucleotides) and number of ambiguous bases (Phred Quality Score higher than 20). Quality trimming was performed. The mean numbers of reads from different groups after trimming ranged from 3.2 million to 8.4 million. The trimmed reads were then mapped to Homo sapiens gene sequences based on Homo sapiens (hg19) mRNA.
Bioconductor edgeR in R package was employed to perform the differential expression analysis. A general linear model was applied on the subjects before and after HCV infection to accommodate the multifactor design of the experiment. The model incorporates the main effect for HCV infection plus interactions with patients and viral clearance, thus allowing us to identify genes differentially expressed in HCV cleared and chronically infected patients before and after HCV infection. To ensure there were sufficient counts for each gene in the test, genes with mean read counts higher than 10 were kept in the analysis. Genes with Benjamini-Hochberg adjusted FDR <0.05 and fold change greater than 1 were considered as significant genes. The significance and function networks of the detected differentially expressed genes were analyzed using Ingenuity Pathway Analysis software. The gene function information was obtained from GeneCards Human Gene Database (www.genecards.org).
Results
Characteristics of study participants
A total of 26 plasma samples from the 13 HCV seroconverters obtained within six months before and after HCV infection were analyzed for plasma transcriptome profiles. All the participants were HIV positive for more than 10 years before HCV infection occurred. Four of the five participants who spontaneously cleared HCV infection had the CC variant of the IL28B gene, while the remaining one had the TT variant. In contrast, half of eight participants who became chronically infected by HCV had the CC variant IL28B gene and the remaining participants had the CT variant. For the 5 individuals with spontaneous HCV clearance, HCV loads were undetectable at the first HCV antibody positive visits. In contrast, the eight participants with chronic HCV infection had a median plasma HCV load of 1.6x107 copies/ml at the first HCV RNA positive visit, with values ranging from 1.22 × 106 to 7 × 107 (Table 1). In the HCV clearance group, the HIV viral load was below 40 copies/ml in three of the five patients and > 71,000 copies/ml in two participants both before and after HCV seroconversion. In the chronically HCV infected group, HIV viral load ranged from 360 to 465,000 copies/ml (mean 125,233 copies/ml) (Table 1).
Plasma mRNA profile of HIV-infected individuals with HCV clearance and those with chronic infection
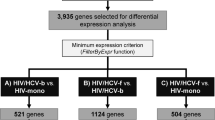
Plasma transcriptome analysis was performed in HCV infected individuals who cleared virus and those who remained chronically infected. A schematic diagram of the approaches for measurement of differentially expressed genes from the patients’ plasma samples collected before and after HCV infection and those cleared HCV and those who remained chronically infected is shown in Fig. 1. A comparison of transcriptomes before HCV infection between the HCV clearance group (TR1B, Fig. 1) and the chronically HCV infected group (TR 2B, Fig. 1) showed that the expression levels of 32 genes were significantly higher (5–563 fold) and expression level of one gene (LL22NC03–63E9.3) was significantly lower in the clearance group (Table 2). To further assess the clustering of subjects within the chronic and clearance groups and across the groups, hierarchical clustering was performed using the differentially expressed genes (Fig. 2a). Four of the eight chronically infected participants clustered together indicating a distinct gene expression pattern whereas the other four samples did not show a similar pattern. On the other hand, three of the five participants in the clearance group clustered indicating similar gene expression and the remaining two subjects had different gene expression. In both clearance and chronic groups, the characteristics of the samples that did not aggregate with the rest of the samples in the respective groups were not different with respect to HIV viral load or IL28B polymorphism. This suggests that HIV viral load and IL28B polymorphism may not have an independent effect on the pattern of the gene expression.
Differentially expressed genes before HCV infection in clearance group compared to chronic group. a The heatmap was generated with the statistical differentially expressed genes between clearance group and chronic group before HCV infection. The numbers on X-axis represent individual subjects. Green stands for the clearance group and blue stands for the chronic group. b Pie chart illustrates the percentage of the DEGs, which encode proteins involved in different functions
Of the 33 genes that were differentially expressed (DEGs) before HCV infection, 19 DEGs have known biological functions (Gene Cards Human Gene Database (http://www.genecards.org/)): six gene products (18%) (ATP6V1G2, LPAR1, GNAI1, CCL3, IL17D, SLC2A6) are related to signal transduction pathways and innate immune response,. eight gene products (24%) (LBR, EARS2, RAB11FIP5, CHST10, PRPSAP1, PCMTD2, MCU, PRDM13) are involved in metabolism and protein trafficking, and five gene products (15%) (ZFHX3, DAZ4, TLX3, HOXD13, NFKBIL1) are RNA/DNA binding proteins involved in gene regulation (Fig. 2b).
To determine the differential gene expression pattern in response to HCV infection, a similar analysis was performed between transcriptomes in the clearance group (TR1A, Fig. 1) and chronic infection group (TR2A, Fig. 1), after HCV infection (Fig. 1). The expression levels of 56 genes were significantly higher, and that of one gene was significantly lower, in TR1A compared to TR2A (Table 3). Hierarchical clustering of DEGs showed that genes from five patients who had chronic infection clustered together, while no such clustering pattern was observed in the clearance group (Fig. 3a).
Differentially expressed genes after HCV infection in clearance group compared to chronic group. a The heatmap was generated with the statistical differentially expressed genes between clearance group and chronic group after HCV infection. The numbers on X-axis represent individual subjects. Green stands for the clearance group and blue stands for the chronic group. b The pie chart illustrates the percentage of the DEGs, which encode proteins involved in different functions
Of the 56 DEGs in the HCV clearance group, 34 are known to encode biologically functional proteins. Of these 34 genes, 14 DEG gene products (25%) (MIXL1, SLC30A3, SFRP5, NLRP13, IRS4, GNAI1, CMTM4, F10, LPAR1, ATP12A, TLX3, ATP6V1G2, NEURL, AP3B1) are known to participate in, or to be related to, signal transduction pathways and innate immune responses, twelve DEG gene products (21%) (LBR, MTMR1, PPP1R3G, PRPSAP1, RAB11FIP5, MANEAL, MCU, ADPRHL1, PRDM13, DPY19L3, SLC51A, CHST10) are involved in metabolism and protein trafficking and eight DEG gene products (14%) (ZFHX3, SOHLH1, TCF15, ZC3H8, ZXDB, TLX3, HOXD13, ZXDA) are RNA/DNA binding proteins involved in gene regulation (Fig. 3b). However, no DEG was identified between plasma samples analyzed before and after HCV infection, regardless whether they were from clearance group (TR1B vs. TR1A) or chronic group (TR 2B vs. TR 2A) (Fig. 1).
Intersection analyses between DEGs identified in the clearance group before HCV infection (DEG-B, Fig. 1) and after HCV infection (DEG-A, Fig. 1) indicate that 12 DEGs were uniquely expressed in clearance group before HCV infection and 36 DEGs were uniquely expressed after HCV infection (Fig. 4). There were 21 DEGs that were common between the two groups. IPA analyses of these unique 12 genes expressed before HCV infection indicate that a number of these genes and their upstream regulators are associated with innate immune response in the clearance group (Fig. 5a and b). Among them 14% are involved in immune cell trafficking, 14% are involved in humoral immune response and 14% are involved in cell mediated immune response suggesting that most of the unique DEGs in the clearance group before infection may play a role in defending against the virus (Fig. 5a). For instance, cytokine (such as IL-1, IL-4, IL-6, IL-8, IL-9, IL-10, TNFR1, interferon) signaling, Toll like receptor (TLR) signaling, MAPK signaling, NF-κB signaling, and communication between innate and adaptive immune cells may be involved (Fig. 5b). Similar analyses with 36 DEGs after HCV infection found that these genes are involved in glucocorticoid receptor signaling, myc mediated apoptosis signaling, axonal guidance signaling, IGF1 signaling, MAPK signaling, p53 signaling, PI3K/AKT signaling, and acute phase response signaling.
Physiological system development and function of unique DEG in clearance group before infection. a Pie chart demonstrates that majority of DEGs seen before HCV infection in the clearance group is clustered in immune mediated pathways. b Predicted interaction networks of mRNAs and associated pathways were identified using the DEGs in clearance compared to chronic group before HCV infection. The interactions between genes were identified using Ingenuity Pathway Analysis
Discussion
Unraveling the mechanism of spontaneous HCV clearance after an acute phase of infection in HIV infected individuals provides valuable information that will be informative for the development of a vaccine against HCV. In a genome-wide association study of HCV infected individuals, SNPs in the region of the IL28B gene, encoding interferon-λ-3, were shown to be associated with the outcome of HCV infection; the CC genotype enhances spontaneous resolution of HCV infection, whereas those with the TT genotype showed a great propensity to develop chronic HCV infection [15, 16]. In the current study we could not detect any significant relationship between CC genotype and HCV clearance, because the CC genotype was found in four of the five clearance subjects and four of the eight chronically HCV infected subjects. The distribution of the genotypes in the infected subjects suggests that other factors beside the IL28B SNPs play a role in the spontaneous HCV clearance in the co-infected individuals [14, 18, 27]. However, the small sample size in our study also could explain our failure to detect an association between IL28B polymorphisms and HCV clearance.
Recent advances in RNA sequencing technology have provided opportunities to comprehensively and quantitatively sequence RNA and examine virus-host interactions at the transcriptional level. Two studies have reported transcriptomes analysis of liver tissues of HCV infected chimpanzees with spontaneous viral clearance or chronic infection outcomes. Nanda et al [18] reported that early induction of the genes associated with cell proliferation and immune activation, especially interleukin binding factor 3 (ILF3) and cytotoxic granule associated RNA binding protein (TIA1), was associated with subsequent HCV clearance in infected chimpanzees. Suet et al [28] reported that cellular genes induced by IFN-γ and those involved in antigen processing/presentation and the adaptive immune response were associated with HCV clearance. These reports suggest that host innate/adaptive immune responses play an important role in HCV clearance. However, studies on the transcriptome of liver tissues from HIV/HCV co-infected humans with different outcomes are very limited. Due to lack of available liver tissues, we focused on determining transcriptomes in plasma from HIV/HCV co-infected subjects with and without HCV clearance, since the quantity and characteristics of RNAs in plasma may reflect concurrent pathogenic changes in host liver tissues.
The availability of sequential plasma samples with defined onset and disease outcomes of HCV infection in highly characterized HIV-infected subjects from the MACS [25] provided us with a unique opportunity to identify, for the first time, the cellular transcripts that may be related to HCV clearance. Application of RNA sequencing technology in plasma has allowed us to determine such differentially expressed transcripts between subjects who cleared HCV and those who remained chronically infected after acute infection. In addition, the analysis of samples collected before and after acquisition of HCV infection provided information on cellular transcripts that may be associated with HCV clearance. Plasma transcriptome analysis of mRNA in subjects who cleared HCV and those who remained chronically HCV infected before and after HCV infection identified 12 DEGs that are uniquely expressed before HCV infection and a number of these genes and their upstream regulators are associated with innate immune response in the clearance group. For instance, IL17D, a member of IL17 family, plays a major regulatory role in host defense and inflammatory diseases [29]. The NFKBIL1 gene encodes a divergent member of the I-kappa-B family of proteins and is involved in the regulation of innate immune response by negatively regulating TLR and interferon regulatory factor (IRF) signaling pathways. In addition, the NFKBIL1 gene has a role in negative regulation of transcriptional activation of NF-kB genes in response to pro-inflammatory stimuli [30]. CCL3, a chemokine, has been shown to increase NK activity [31]. This goes along with observation that subjects who resolved HCV infection had a higher frequency of HCV-specific interferon-gamma-producing T-cells (P = 0.017) and cytotoxic NK-cells (P = 0.005), compared to patients who became chronically infected [27]. In addition, in HCV infected patients treated with interferon-α and ribavirin, increases in MIP-1α, MIP-1β and RANTES levels after 24 h of treatment were exclusively observed in the group that showed a sustained virological response, suggesting an antiviral role of CCL3 [32]. Furthermore, IPA analysis suggests that cytokine signaling (such as IL-1, IL-4, IL-6, IL-8, IL-9, IL-10, TNFR1, interferon), TLR signaling, MAPK signaling, NF-κB signaling, communication between innate and adaptive immune cells, and ELF3 could be involved in HCV clearance.
Conclusions
Plasma RNA sequencing has identified the transcripts that are significantly associated with HCV clearance and are found to be expressed prior to infection. Some of the transcripts are involved in innate immune function. These results suggest that subjects who spontaneously clear HCV may have the transcripts that modulate forms of immunity that confer resistance to chronic infection with HCV. Further studies with longitudinal samples from HCV infected patients who cleared HCV infection, and those who became chronically infected may provide more definitive information about the nature of these inherited traits.
Abbreviations
- cDNA:
-
Complementary DNA
- DEG-A:
-
Differentially expressed genes after HCV infection
- DEG-B:
-
Differentially expressed genes before HCV infection
- DEGs:
-
Differentially expressed genes
- FDR:
-
False Discovery Rate
- HCV:
-
Hepatitis C virus
- HIV:
-
Human immunodeficiency virus
- IL28B:
-
Polymorphisms in interleukin 28B
- ILF3:
-
Interleukin binding factor 3
- MACS:
-
The Multicenter AIDS Cohort Study
- TLR:
-
Toll like receptor
- TR1A:
-
HCV clearance group after HCV infection
- TR1B:
-
HCV clearance group before HCV infection
- TR2A:
-
Chronically HCV infected group after HCV infection
- TR2B:
-
Chronically HCV infected group before HCV infection
References
El-Serag HB. Hepatocellular carcinoma and hepatitis C in the United States. Hepatology. 2002;36(5 Suppl 1):S74–83.
Sherman KE, Rouster SD, Chung RT, Rajicic N. Hepatitis C Virus prevalence among patients infected with Human Immunodeficiency Virus: a cross-sectional analysis of the US adult AIDS Clinical Trials Group. Clin Infect Dis. 2002;34(6):831–7.
Ghosn J, Deveau C, Goujard C, Garrigue I, Saichi N, Galimand J, Nagy Z, Rouzioux C, Meyer L, Chaix ML. Increase in hepatitis C virus incidence in HIV-1-infected patients followed up since primary infection. Sex Transm Infect. 2006;82(6):458–60.
Urbanus AT, van de Laar TJ, Stolte IG, Schinkel J, Heijman T, Coutinho RA, Prins M. Hepatitis C virus infections among HIV-infected men who have sex with men: an expanding epidemic. AIDS. 2009;23(12):F1–7.
Micallef JM, Kaldor JM, Dore GJ. Spontaneous viral clearance following acute hepatitis C infection: a systematic review of longitudinal studies. J Viral Hepat. 2006;13(1):34–41.
Weber R, Sabin CA, Friis-Moller N, Reiss P, El-Sadr WM, Kirk O, Dabis F, Law MG, Pradier C, De Wit S, et al. Liver-related deaths in persons infected with the human immunodeficiency virus: the D:A:D study. Arch Intern Med. 2006;166(15):1632–41.
Lewden C, Salmon D, Morlat P, Bevilacqua S, Jougla E, Bonnet F, Heripret L, Costagliola D, May T, Chene G, et al. Causes of death among human immunodeficiency virus (HIV)-infected adults in the era of potent antiretroviral therapy: emerging role of hepatitis and cancers, persistent role of AIDS. Int J Epidemiol. 2005;34(1):121–30.
Sahasrabuddhe VV, Shiels MS, McGlynn KA, Engels EA. The risk of hepatocellular carcinoma among individuals with acquired immunodeficiency syndrome in the United States. Cancer. 2012;118(24):6226–33.
Seaberg EC, Witt MD, Jacobson LP, Detels R, Rinaldo CR, Margolick JB, Young S, Phair JP, Thio CL. Spontaneous Clearance of the Hepatitis C Virus Among Men Who Have Sex With Men. Clin Infect Dis. 2015;61(9):1381–8.
Witt MD, Seaberg EC, Darilay A, Young S, Badri S, Rinaldo CR, Jacobson LP, Detels R, Thio CL. Incident hepatitis C virus infection in men who have sex with men: a prospective cohort analysis, 1984–2011. Clin Infect Dis. 2013;57(1):77–84.
Alric L, Fort M, Izopet J, Vinel JP, Bureau C, Sandre K, Charlet JP, Beraud M, Abbal M, Duffaut M. Study of host- and virus-related factors associated with spontaneous hepatitis C virus clearance. Tissue Antigens. 2000;56(2):154–8.
Balagopal A, Thomas DL, Thio CL. IL28B and the control of hepatitis C virus infection. Gastroenterology. 2010;139(6):1865–76.
Mosbruger TL, Duggal P, Goedert JJ, Kirk GD, Hoots WK, Tobler LH, Busch M, Peters MG, Rosen HR, Thomas DL, et al. Large-scale candidate gene analysis of spontaneous clearance of hepatitis C virus. J Infect Dis. 2010;201(9):1371–80.
Beinhardt S, Aberle JH, Strasser M, Dulic-Lakovic E, Maieron A, Kreil A, Rutter K, Staettermayer AF, Datz C, Scherzer TM, et al. Serum level of IP-10 increases predictive value of IL28B polymorphisms for spontaneous clearance of acute HCV infection. Gastroenterology. 2012;142(1):78–85–e72.
Bibert S, Roger T, Calandra T, Bochud M, Cerny A, Semmo N, Duong FH, Gerlach T, Malinverni R, Moradpour D, et al. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med. 2013;210(6):1109–16.
Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461(7265):798–801.
Tillmann HL, Thompson AJ, Patel K, Wiese M, Tenckhoff H, Nischalke HD, Lokhnygina Y, Kullig U, Gobel U, Capka E, et al. A polymorphism near IL28B is associated with spontaneous clearance of acute hepatitis C virus and jaundice. Gastroenterology. 2010;139(5):1586–92. 1592 e1581.
Nanda S, Havert MB, Calderon GM, Thomson M, Jacobson C, Kastner D, Liang TJ. Hepatic transcriptome analysis of hepatitis C virus infection in chimpanzees defines unique gene expression patterns associated with viral clearance. PLoS One. 2008;3(10):e3442.
Martin MP, Carrington M. Immunogenetics of HIV disease. Immunol Rev. 2013;254(1):245–64.
Chen YJ, Zhu JM, Wu H, Fan J, Zhou J, Hu J, Yu Q, Liu TT, Yang L, Wu CL, et al. Circulating microRNAs as a Fingerprint for Liver Cirrhosis. PLoS One. 2013;8(6):e66577.
Oloomi M, Bouzari S, Mohagheghi MA, Khodayaran-Tehrani H. Molecular markers in peripheral blood of Iranian women with breast cancer. Cancer Microenviron. 2013;6(1):109–16.
Shen J, Wang H, Wei J, Yu L, Xie L, Qian X, Zou Z, Liu B, Guan W. Thymidylate synthase mRNA levels in plasma and tumor as potential predictive biomarkers for raltitrexed sensitivity in gastric cancer. Int J Cancer. 2012;131(6):E938–945.
Tzimagiorgis G, Michailidou EZ, Kritis A, Markopoulos AK, Kouidou S. Recovering circulating extracellular or cell-free RNA from bodily fluids. Cancer Epidemiol. 2011;35(6):580–9.
Wang W, Zhang X, Xu Y, Di Bisceglie AM, Fan X. Viral categorization and discovery in human circulation by transcriptome sequencing. Biochem Biophys Res Commun. 2013;436(3):525–9.
Kaslow RA, Ostrow DG, Detels R, Phair JP, Polk BF, Rinaldo Jr CR. The Multicenter AIDS Cohort Study: rationale, organization, and selected characteristics of the participants. Am J Epidemiol. 1987;126(2):310–8.
Soto-Rivera J, Patterson BK, Chen Y, Shen C, Ratner D, Ding M, Tumne A, Gupta P. Study of HIV-1 transmission across cervical mucosa to tonsil tissue cells using an organ culture. Am J Reprod Immunol. 2013;69(1):52–63.
Riva A, Laird M, Casrouge A, Ambrozaitis A, Williams R, Naoumov NV, Albert ML, Chokshi S. Truncated CXCL10 is associated with failure to achieve spontaneous clearance of acute hepatitis C infection. Hepatology. 2014;60(2):487–96.
Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, Wieland S, Bukh J, Purcell RH, Schultz PG, et al. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci U S A. 2002;99(24):15669–74.
O’Sullivan T, Saddawi-Konefka R, Gross E, Tran M, Mayfield SP, Ikeda H, Bui JD. Interleukin-17D mediates tumor rejection through recruitment of natural killer cells. Cell Rep. 2014;7(4):989–98.
Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat Immunol. 2011;12(8):695–708.
Robertson MJ. Role of chemokines in the biology of natural killer cells. J Leukoc Biol. 2002;71(2):173–83.
Zhang Y, Guo D, Zhao Y, Chen X, Ma L, Jin Y, Yan H, Wu H, Wei L, Dong T, et al. The effect of cytokine profiles on the viral response to re-treatment in antiviral-experienced patients with chronic hepatitis C virus infection. Antiviral Res. 2011;92(2):247–54.
Acknowledgements
Data in this article were collected by the Multicenter AIDS Cohort Study (MACS) with centers (principal investigators) at The Johns Hopkins Bloomberg School of Public Health (Joseph B. Margolick, Lisa P. Jacobson), Howard Brown Health Center, Feinberg School of Medicine, Northwestern University, and Cook County Bureau of Health Services (John P. Phair, Steven M. Wolinsky), University of California, Los Angeles (Roger Detels), and University of Pittsburgh (Charles R. Rinaldo).
Funding
The MACS is funded by the National Institute of Allergy and Infectious Diseases, with additional supplemental funding from the National Cancer Institute (UO1- AI-35042, UL1-RR025005, UO1-AI-35043, UO1-AI-35039, UO1-AI-35040, UO1-AI-35041). Website located at https://statepi.jhsph.edu/macs/macs.html.
Availability of data and materials
Data are held by the Center for Analysis and Management of the Multicenter AIDS Cohort Study (CAMACS). For access to the MACS data, please contact CAMACS at https://statepi.jhsph.edu/macs/macs.html.
Authors’ contributions
YC and PG conceived of the study. CS and DG provided assistance with experimental design and sample section. CR, ES, JM, VS, and OM contributed samples for the study. YC, CS, AA, SK and MD designed and performed the laboratory experiments as well as analyzed the results. YC and PG drafted the original manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study was approved by the Institutional Review Board (IRB)/ethical standards committee in University of Pittsburgh, University of California, Los Angeles, Northwestern University and John Hopkins University. Written informed consent was obtained from all subjects participating in the study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Chen, Y., Shen, C., Guha, D. et al. Identification of the transcripts associated with spontaneous HCV clearance in individuals co-infected with HIV and HCV. BMC Infect Dis 16, 693 (2016). https://doi.org/10.1186/s12879-016-2044-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-016-2044-7