Abstract
Background
Gastric cancer develops even in Helicobacter pylori(H. pylori)-uninfected patients and its typical histological feature is signet ring cell carcinoma (SRCC) within the mucosal layer. However, the biological characteristics of SRCC remain unclear. We aimed to clarify the pathological and genetic features of SRCC in H. pylori-uninfected patients.
Methods
Seventeen H. pylori-uninfected patients with mucosal SRCCs were enrolled and their clinicopathological characteristics were compared with those of H. pylori-infected patients with mucosal SRCCs. Seven SRCCs without H. pylori-infected, including two invasive SRCCs, and seven H. pylori-infected SRCCs were subjected to a genetic analysis using next-generation sequencing.
Results
H. pylori-uninfected patients with mucosal SRCCs revealed male dominancy and a significantly higher prevalence of smokers among them as compared with the H. pylori-infected patients with SRCC. A CDH1 mutation (frame shift indel) was detected in one H. pylori-uninfected cancer not only in the mucosal SRCC but also in the invasive portion. A TP53 mutation was detected in one SRCC without H. pylori-infected. In the control group, ARID1A and TP53 mutations were detected in one SRCC each. The C to A mutation, which is a characteristic smoking-induced mutation, was not found in any of the samples.
Conclusions
Some SRCCs in H. pylori-uninfected patients may have a malignant potential similar to that of SRCCs in H. pylori-infected patients. Smoking may not be the main carcinogenic factor for the development of SRCCs among the H. pylori-uninfected patients.
Similar content being viewed by others
Background
It is well recognized that Helicobacter pylori (H. pylori)-infected is the main cause of gastric cancer development [1]. In Japan, we previously reported that over 99% of patients with gastric cancer had H. pylori-infected [2], and another study reported a similar result [3]. However, gastric cancer may develop even in H. pylori-uninfected patients and its typical histological feature is signet ring cell carcinoma (SRCC) within the mucosal layer [4]. Endoscopically, SRCCs can be easily recognized as whitish depressed or flat lesions, and their prevalence and clinical importance have recently been increasing [5].
Previous reports have suggested that SRCCs in H. pylori-uninfected patients show a lower proliferative activity, few extensive spread, and slower progression, compared with SRCCs with H. pylori-infected patients [6]. The features of SRCCs in H. pylori-uninfected patients resemble those in patients with hereditary diffuse gastric cancer (HDGC), which is caused by a germline mutation of CDH1 [7]. In patients with HDGC, a large number of SRCCs can be detected; however, most of them show little tendency to invade into the submucosal layer. Therefore, it is controversial whether SRCCs in H. pylori-uninfected patients can invade into the submucosal layer. In addition, the pathogenesis of SRCCs in H. pylori-uninfected patients remains uncertain. Recently, Horiuchi et al. reported the implication of smoking in the disease pathogenesis of these patients, noting that their findings should be verified by other studies [8].
In the present study, we compared the clinical features of SRCCs between patients with and without H. pylori-infected. Moreover, we investigated the genomic characteristics of these lesions to identify their biological characteristics and assess whether smoking may be involved in their pathogenesis.
Methods
Patients
We enrolled 19 consecutive patients with SRCC (17 with mucosal cancer and 2 with invasive cancer) without H. pylori-infected who were diagnosed in Hiroshima University Hospital from 1998 to 2017. All the patients received endoscopic or surgical resection. The entire tumor was cut into parallel 2–4 mm-thick sections for investigation. Histological evaluation was performed according to the Japanese gastric cancer treatment guidelines [9]. Clinicopathological data was retrospectively reviewed from clinical records. The definition of H. pylori-uninfected subjects was judged by our criteria as described in a previous publication [2].
A total of 34 patients with H. pylori-infected SRCC diagnosed in the same period were enrolled as controls. We compared the clinical characteristics (sex, age, location, size, blood type, family history of gastric cancer, alcohol, smoking, and Brinkman Index) between the two groups (17 patients H. pylori-uninfected mucosal SRCC and 34 patients controls). Furthermore, we randomly selected seven patients who were habitual smokers among both the H. pylori-uninfected and -infected SRCCs, for next-generation sequencing DNA analysis. This retrospective study was approved by the institutional review board and the ethics committees of Hiroshima University (No. E-1096) and was performed in accordance with the Helsinki Declaration and its later amendments.
Tissue capture and DNA extraction
In the experiments with next-generation sequencing, we randomly selected seven patients who were habitual smokers among the H. pylori-uninfected SRCCs.
In addition, two DNA samples (one from an SRCC within the mucosal layer and one from a deeper PDA lesion) were extracted from two cases with invasive SRCC without H. pylori-infected (Case 5 and 6).
We examined 10 diffuse-type gastric cancer associated genes (CDH1, TP53, ARID1A, KRAS, PIK3CA, ERBB3, FBXW7, TGFBR1, RHOA, and MAK2K1) referring to previous paper [10].
In the control group, seven samples were collected from only mucosal SRCC with H. pylori-infected. Therefore, in total, nine and seven DNA samples were extracted from the H. pylori-uninfected and -infected patients, respectively.
Pathologic tumor tissues were dissected from ten 10-μm-thick slides made from formalin-fixed paraffin embedded (FFPE) specimens, which were deparaffinized, stained, and dehydrated using the Arcturus® Paradise® PLUS Reagent System (Thermo Fisher Scientific, Waltham, MA, USA). Dissections were performed using a laser capture microdissection system (LMD 6500, Leica, Wetzlar, Germany), in accordance with the pathogenesis diagnosis. The DNA was extracted from these tissues using the GeneRead DNA FFPE kit (Qiagen, Valencia, CA, USA), and concentrations were determined using a Qubit® 1.0 fluorometer (Life Technologies, Carlsbad, CA, NY, USA). The quantity and quality of the FFPE-derived DNA samples were checked by calculating the normalized DNA integrity scores (ΔΔCq) via quantitative polymerase chain reaction (PCR) analysis using the Agilent NGS FFPE QC kit (Agilent, Santa Clara, CA, USA).
Target enrichment and next-generation sequencing
DNA extracted from tumors was fragmented into 150–200 bp portions by sonication using an S2 sonicator (6 min, 10% duty, intensity = 5, 200 cycles/burst; Covaris, Woburn, MA, USA) and used for library construction according to the manufacturer’s instructions. In all cases, 10 ng of DNA was prepared for sequencing. The exons of ten oncogenes (CDH1, TP53, ARID1A, KRAS, PIK3CA, ERBB3, FBXW7, TGFBR1, RHOA, and MAK2K1) were enriched using the SureSelectXT HS Custom panel (Agilent). The resulting pooled libraries were quality control-checked via the High Sensitivity D1000 ScreenTape system using the 2200 TapeStation instrument (Agilent). Sequencing was performed with paired-end reads via the HiSeq 2500 platform (Illumina, San Diego, CA, USA).
Variant detection
Sequencing reads were aligned to the hg19/GRCh37 reference sequence and analyzed using SureCall 4.0.1 (Agilent). PCR duplicates were removed and low-frequency mutations in template DNA molecules were detected using the molecular barcode system [11]. To identify variants in tumor samples, single sample analysis in SureCall 4.0.1 was used. The called variants were considered germline mutations if they were found in the dbSNP 137 or TogoVar (https://togovar.biosciencedbc.jp) databases. The remaining mutations in cancerous tissues were considered to be candidate cancer-specific mutations. To reduce the false positive rate, we set the cutoff values for somatic mutation in cancerous tissues as follows: variant score > 0.3; minimum quality for base > 30; variant call quality > 100; allele frequency > 0.1; and number of reads supporting the variant allele > 3 (Supporting file 1).
Statistical analysis
All clinicopathological features were analyzed using the Chi-square test or Fisher’s exact test to compare categorical data and Student’s t test or Wilcoxon rank-sum test to compare continuous data. A p-value of < 0.05 was considered significant. All statistical analyses were performed using JMP® software (SAS International, Cary, NC. USA).
Results
Clinical features in H. pylori-uninfected and -infected SRCCs
First, we compared the clinicopathological features between 17 H. pylori-uninfected and -infected patients with SRCCs located within the mucosal layer. As shown in Table 1, the H. pylori-uninfected patients with mucosal SRCCs revealed male dominancy and a slightly younger age than the H. pylori-infected patients. The SRCCs in the H. pylori-uninfected patients tended to be located in the lower third of the stomach and its tumor size were significantly smaller than those in the H. pylori-infected patients. Although no difference was detected in blood type, family history of gastric cancer, or alcohol consumption.
Genomic characteristics of SRCCs
DNA analysis by next-generation sequencing was performed in 16 samples from 14 patients (seven patients from each group) as shown in Table 2.
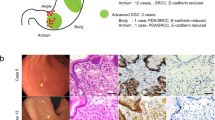
A single gene mutation was detected in each of five samples (Table 3, Fig. 1). A CDH1 mutation (frame shift indel) was detected only in one H. pylori-uninfected cancer lesion (case 6) (Fig. 1). This mutation was found not only in the mucosal SRCC but also in the invasive PDA of that patient (Figs. 2 and 3). A TP53 mutation was detected in one SRCC without H. pylori-infected (case 4). In the H. pylori-infected control group, mutations in ARID1A (case 11) and TP53 (case 12) were detected in SRCCs (Fig. 1). No mutation in CDH1 was found in the control group. On the other hand, we did not find somatic mutation in 7 genes (KRAS, PIK3CA, ERBB3, FBXW7, TGFBR1, RHOA, and MAK2K1) which associated somatic genomic alterations associated with the unique characteristics of sporadic diffuse gastric cancers [10]. (Fig. 1).
Mutational landscapes of Seven SRCCs without H. pylori-infected, and seven H. pylori-infected SRCCs. Gastric signet ring cell carcinoma (SRCCs) tissues, paired non-cancerous tissues, from Seven SRCCs without H. pylori-infected, and seven H. pylori-infected SRCCs were subjected to the targeted panel for 10 genes. The Upper bar-graph shows the number of somatic mutations per sample. The Lower panel shows the type of mutation of each gene. Genes reported SRCC-associated genes in previous paper. First line shows H. pylori-infected, second line shows smoking status, and from 3th to 12th line shows the type of mutation of each gene. Blue panels shows H. pylori-infected cases, brown panels shows present or current smoker, light-brown panels shows non-smoker, red panels shows frameshift indels or nonsense mutations, and yellow panels shows missence mutations
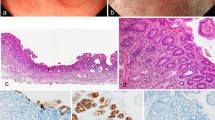
Histological features of case 6. A depressed lesion was found in the center of the section (a) and the low magnitude feature [red box in (a)] was demonstrated in (b). The histologic features of a higher magnitude mucosal signet ring cell carcinoma (yellow box) and a poorly differentiated adenocarcinoma in the deep invasive portion (blue box) are shown in (c) and (d), respectively (hematoxylin and eosin staining)
The association between smoking status and, clinical and genomic characterization in SRCCs
As a result of comparison for smoking status in H. pylori-uninfected and -infected patients with SRCCs, the prevalence of smokers was significantly higher among the pylori-uninfected patients (p = 0.02). Although, the Brinkman index was not different between the two groups. Moreover, the C to A mutation, which is a characteristic smoking-induced mutation, was not found in any of the 17 H. pylori-uninfected mucosal SRCC.
Discussion
In the present study, we examined the clinical and genetic characteristics of SRCCs in patients without H. pylori-infected. SRCCs in patients without H. pylori-infected showed different features compared with those in patients with H. pylori-infected. Clinically, the H. pylori-uninfected patients with mucosal SRCCs showed male dominance and the tumor tissue tended to be located in the lower third of the stomach. These observations imply that H. pylori-uninfected SRCCs have distinct biological characteristics and probably have a different carcinogenetic pathway as compared with H. pylori-infected SRCCs.
Particular focus should be placed on the genetic alterations detected in SRCCs without H. pylori-infected. Generally, diffuse-type cancers including SRCC are classified into the “genomically stable group” according to the four-subtype classification [12]. Under these conditions, CDH1 have frequently been detected in SRCCs in patients with HDGC [13]. Moreover, E-cadherin, which is coded by CDH1 gene, regulates signaling pathways leads to increase in cell proliferation, decrease in cell apoptosis followed by gastric cancer development [14]. However, in our study, the prevalence of mutations in CDH1 was extremely low in SRCCs without H. pylori- infected. This suggests that the carcinogenic pathway of SRCCs without H. pylori- infected differs from that of SRCCs in patients with HDGC. The SRCC lesions found in H. pylori-uninfected patients seem to reflect not only a defect in cell-cell adhesion but also other neoplastic alterations.
Mutations in TP53, which are known as a representative driver mutation in gastric cancer carcinogenesis, were detected in SRCCs without H. pylori-infected as well as those with H. pylori-infected. This also strongly suggests that SRCCs in H. pylori-uninfected patients may be true neoplastic lesions. Previous reports have demonstrated that SRCC tissue gains an invasive ability after the mutation of TP53 followed by conversion to PDA [15]. Accordingly, at least in a part of H. pylori-uninfected SRCC is a true neoplasm, and clinical features of SRCC without H. pylori-infected may be different from that in HDGC. In the present study, we demonstrated the presence of similar genetic alterations between mucosal SRCC and invasive PDA in a H. pylori-uninfected patient, suggesting the progression of SRCC to invasive PDA. Indeed, we have diagnosed two cases with invasive SRCC without H. pylori-infected (paper under submission). SRCC without H. pylori-infected may have a malignant potential to invade into the submucosal layer and should be treated as a diffuse-type gastric cancer as described in the guidelines for treatment [16].
Notably, it should be emphasized that the prevalence of smokers was higher among the H. pylori-uninfected patients with SRCCs than among the H. pylori-infected patients. This finding is compatible with the previous report by Horiuchi et al. [8]. However, in our study concerning the genetic alterations in SRCC tissue samples, we could not find any C to A mutations, which are recognized as smoking-induced alterations, despite including samples from some heavy smokers in our analysis [17, 18]. Smoking may be a confounding factor as to the pathogenesis of SRCCs without H. pylori-infected.
In addition to genetic mutations, gastric carcinogenesis may also be influenced by epigenomic alterations including gene methylation. Epstein–Barr virus (EB virus) infection, which induces several epigenetic alterations in host genes, may be another carcinogenetic factor besides H. pylori-infected [19]. However, it is supposed that H. pylori-infected is essential for EB virus-induced carcinogenesis [20]. The pathogenesis of SRCCs without H. pylori-infected remains unclear and should be clarified in future research.
This study has several limitations. The first limitation is the limited number of patients. To accurately evaluate the prevalence of any given genetic mutation, a large number of cases should be examined. In addition, we examined only ten genes in our cancer panel and we could not cover their complete exon sequences. In clinical practice, SRCCs without H. pylori-infected are diagnosed when the lesions measure about 10 mm in diameter; therefore, it is difficult to extract high quality DNA from SRCC tissue. The second limitation is that DNA samples from control lymphocytes were not analyzed. Therefore, we excluded disease-specific germline mutations and Japanese healthy germline mutations when searching the dbSNP 137 and TogoVar databases.
Conclusions
Our findings indicate that SRCCs without H. pylori should be treated as having a malignant potential similar to that for SRCCs occurring in the presence of H. pylori-infected. Smoking may not be the main carcinogenic factor for the development of SRCCs among the H. pylori-uninfected patients.
Availability of data and materials
The datasets used and analyzed during the current study will be available from the corresponding author on reasonable request.
Abbreviations
- SRCC:
-
Signet ring cell carcinoma
- H. pylori :
-
Helicobacter pylori
- HDGC:
-
Hereditary diffuse gastric cancer
- PDA:
-
Poorly differentiated adenocarcinoma
- FFPE:
-
Formalin-fixed paraffin embedded
- EB virus:
-
Epstein–Barr virus
References
IARC Working Group Reports Volume 8. Helicobacter pylori Eradication as a Strategy for Preventing Gastric Cancer.
Matsuo T, Ito M, Takata S, Tanaka S, Yoshihara M, Chayama K. Low prevalence of Helicobacter pylori-negative gastric cancer among Japanese. Helicobacter. 2011;16:415–9.
Ono S, Kato M, Suzuki M, Ishigaki S, Takahashi M, Haneda M, et al. Frequency of Helicobacter pylori-negative gastric cancer and gastric mucosal atrophy in a Japanese endoscopic submucosal dissection series including histological, endoscopic and serological atrophy. Digestion. 2012;86:59–65.
Kiso M, Yoshihara M, Ito M, Inoue K, Kato K, Nakajima S, et al. Characteristics of gastric cancer in negative test of serum anti-Helicobacter pylori antibody and pepsinogen test: a multicenter study. Gastric Cancer. 2017;20:764–71.
Yamamoto Y, Fujisaki J, Omae M, Hirasawa T, Igarashi M. Helicobacter pylori-negative gastric cancer: characteristics and endoscopic findings. Dig Endosc. 2015;27:551–61.
Horiuchi Y, Fujisaki J, Yamamoto N, Shimizu T, Miyamoto Y, Tomida H, et al. Biological behavior of the intramucosal Helicobacter pylori-negative undifferentiated-type early gastric cancer: comparison with Helicobacter pylori-positive early gastric cancer. Gastric Cancer. 2016;19:160–5.
Sugimoto S, Komatsu H, Morohoshi Y, Kanai T. Recognition of and recent issues in hereditary diffuse gastric cancer. J Gastroenterol. 2015;50:831–43.
Horiuchi Y, Fujisaki J, Ishizuka N, Omae M, Ishiyama A, Yoshio T, et al. Study on clinical factors involved in Helicobacter pylori-uninfected, undifferentiated-type early gastric cancer. Digestion. 2017;96:213–9.
Japanese Gastric Cancer Association. Japanese gastric cancer treatment guidelines 2014 (ver. 4). Gastric Cancer. 2017;20:1–19.
Cho SY, Park JW, Liu Y, Park YS, Kim JH, Yang H, et al. Sporadic early-onset diffuse gastric cancers have high frequency of somatic CDH1 alterations, but low frequency of somatic RHOA mutations compared with late-onset cancers. Gastroenterology. 2017;153:536–49.
Peng Q, Vijaya Satya R, Lewis M. BMC Genomics. 2015;16:589 https://www.agilent.com/cs/library/brochures/5991-8014EN.pdf.
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9.
Hansford S, Kaurah P, Li-Chang H, Woo M, Senz J, Pinheiro H, et al. Hereditary diffuse gastric cancer syndrome: CDH1 mutations and beyond. JAMA Oncol. 2015;1:23–32.
Liu X, Chu KM. E-cadherin and gastric cancer: cause, consequence, and applications. Biomed Res Int. 2014;2014:637308.
Shimada S, Mimata A, Sekine M, Mogushi K, Akiyama Y, Fukamachi H, et al. Synergistic tumour suppressor activity of E-cadherin and p53 in a conditional mouse model for metastatic diffuse-type gastric cancer. Gut. 2012;61:344–53.
Ono H, Yao K, Fujishiro M, Oda I, Nimura S, Yahagi N, et al. Guidelines for endoscopic submucosal dissection and endoscopic mucosal resection for early gastric cancer. Dig Endosc. 2016;28:3–15.
Smith LE, Denissenko MF, Bennett WP, Li H, Amin S, Tang M, Pfeifer GP. Targeting of lung cancer mutational hotspots by polycyclic aromatic hydrocarbons. J Natl Cancer Inst. 2000;92:803–11.
Le Calvez F, Mukeria A, Hunt JD, Kelm O, Hung RJ, Tanière P, et al. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former, and current smokers. Cancer Res. 2005;65:5076–83.
Saju P, Murata-Kamiya N, Hayashi T, Senda Y, Nagase L, Noda S, et al. Host SHP1 phosphatase antagonizes Helicobacter pylori CagA and can be downregulated by Epstein-Barr virus. Nat Microbiol. 2016;1:16026.
Nishikawa J, Iizasa H, Yoshiyama H, Nakamura M, Saito M, Sasaki S, et al. The role of epigenetic regulation in Epstein-Barr virus-associated gastric cancer. Int J Mol Sci. 2017;18:1606.
Acknowledgements
Quality check by Tapestation (Agilent, Santa Clara, CA, USA) was supported by the Program of the network-type Joint Usaga/Research Center for Radiation Disaster Medical Science of Hiroshima University, Nagasaki University, and Fukushima Medical University. Next-generation sequencing was carried out at the Analysis Center of Life Science, Hiroshima University.
Funding
This study was funded by the Tsuchiya Memorial Medical Foundation (grant number 33–3). This found was acquired by Tomoyuki Boda. The funding body had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
Conception and design of the study; Y.U., M.I., Analysis and interpretation of data; M.K., Y.U., K.M., Collection and assembly of data; K.M., T.B., T.K., K.H., N.Y., N.N., M.A., Y.H., S.O., S.T., Drafting of the article; M.K., Y.U., M.I., Critical revision of the article for important intellectual content; M.K., Final approval of the article; T. B. and K.C. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All procedures were completed in accordance with the Declara- tion of Helsinki of 1964 and later versions. The Ethics Committee of Hiroshima University Hospital approved our study (E-1096). In- formed consent was obtained from all patients by opt-out in our department website.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kiso, M., Urabe, Y., Ito, M. et al. Clinical and genomic characteristics of mucosal signet-ring cell carcinoma in Helicobacter pylori-uninfected stomach. BMC Gastroenterol 20, 243 (2020). https://doi.org/10.1186/s12876-020-01387-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12876-020-01387-9