Abstract
Background
Sugar content is an important determinant of fruit sweetness, but details on the complex molecular mechanism underlying fruit sugar accumulation remain scarce. Here, we report the role of sucrose transporter (SUT) family in regulating fruit sugar accumulation in apple.
Results
Gene-tagged markers were developed to conduct candidate gene-based association study, and an SUT4 member MdSUT4.1 was found to be significantly associated with fruit sugar accumulation. MdSUT4.1 encodes a tonoplast localized protein and its expression level had a negative correlation with fruit sugar content. Overexpression of MdSUT4.1 in strawberry and apple callus had an overall negative impact on sugar accumulation, suggesting that it functions to remobilize sugar out of the vacuole. In addition, MdSUT4.1 is located on chromosomal region harboring a previously reported QTL for sugar content, suggesting that it is a candidate gene for fruit sugar accumulation in apple.
Conclusions
MdSUT4.1 is involved in the regulation of fruit sugar accumulation in apple. This study is not only helpful for understanding the complex mechanism of fruit sugar accumulation, but it also provides molecular tools for genetic improvement of fruit quality in breeding programs of apple.
Similar content being viewed by others
Background
Sucrose is a major product of photosynthesis in plants, and the most common form of carbohydrate transported through the phloem from source leaves to sink organs such as roots and fruits. Extensive research has been conducted in the last three decades on sucrose transport in plants and sucrose transporters (SUTs) are deemed to be essential for the export and efficient movement of sucrose from source to sink [1]. SUTs contain the typical 12 transmembrane (TM) α-helices of the major facilitator superfamily (MFS) proteins [2,3,4]. Most SUTs are characterized as energy-dependent proton sucrose/H+ symporters that utilize the proton motive force across the membrane to transport sucrose against its concentration gradient [5, 6].
The first SUT gene was isolated from spinach by functional complementation in yeast [7]. Later, integrative whole-genome sequence analysis reveals that SUTs belong to a small gene family, with nine and five members in Arabidopsis thaliana [8] and Oryza sativa [9], respectively. Phylogenetic analysis showed that the SUT family could be divided into three subfamilies, SUTI, SUTII, and SUTIII, with the SUTI subfamily present exclusively in eudicots [1, 10,11,12]. The SUTII subfamily are further divided into two clades SUTIIA and SUTIIB, with the latter being monocot specific [11, 12]. SUTIIA represents an ancestral form of the SUTII subfamily in angiosperms and has a longer central cytoplasmic loop than SUTIIB [12, 13]. The SUTI subfamily members are exclusively located in the plasma membrane and are responsible for loading sucrose into the phloem or for uptake of sucrose into cells of sink tissues [14,15,16,17,18,19,20,21]. While most SUTII members are located in the plasma membrane where they function as sugar sensors, recent studies have shown that MdSUT2.2 reside in the vacuolar membrane, and its overexpression leads to increased sucrose accumulation in the vacuole [22, 23]. Members of the SUTIII subfamily have previously been associated with vacuolar membrane where they are involved in sucrose efflux from the vacuole into the cytosol [24,25,26,27,28,29]. However, some SUTIII members, such as Arabidopsis AtSUT4 [30], barley HvSUT2 [31], Lotus japonicus LiSUT4 [25], tobacco NtSUT4 and potato StSUT4 [32], and apple MdSUT1 [33], are located in the plasma membrane. It is worth noting that monocots and eudicots utilize different types of SUTs to load sucrose into the phloem. The SUT1 subfamily that has the highest affinity towards the substrate sucrose is crucial for efficient phloem loading of sucrose in eudicots, whilst monocots utilize the high affinity SUT2 transporters [14, 17, 34, 35]. In addition, SUT4 has a lower affinity for sucrose relative to SUT1 and SUT2 [30].
In addition to their association with sucrose transport, SUT genes also play essential roles in a variety of developmental and physiological processes in plants. For example, the SUT1 gene subfamily participates in regulation of vegetative growth [17], pollen development [36, 37], flowering date [38], fruit and seed development [39, 40], anthocyanin accumulation [41] and tuber yield [42]. The SUT2 gene subfamily was initially described as sugar sensor [13, 43]. However, this putative sucrose-sensing function was subsequently questioned [2], and increasing evidence shows that SUT2 genes function as sugar transporter and contribute to grain yield enhancement [44], reproductive organ development [31] and resistance to abiotic stress [23].
Sugar accumulation is strongly associated with fruit yield and quality. Since sugar transporter genes play important roles in sugar accumulation [45, 46], increasing attention has been paid to investigate SUT gene family over the past two decades in a wide range of fruit species, such as grapevine [47,48,49], sweet orange [50], peach [29, 51], pear [52] and apple [53]. A SUT2 gene, i.e. MdSUT2.2 in apple, has been recently proven to regulate fruit sugar accumulation in apple [22, 23, 54]. Several SUT4 genes, including citrus CsSUT4 [50], grapevine VvSUC11 [55], apple MdSUT1 [33] and jujube ZjSUT4 [56], were found to be potentially associated with fruit sugar accumulation. However, the role of SUT4 gene family in fruit sugar accumulation still remains elusive.
Apple (Malus x domestica Borkh.), a member of the family Rosaceae, is an economically important fruit crop worldwide. It is a diploid but has an autopolyploid origin, with a basic chromosome number of x = 17 and a haploid genome size of approximately 750 Mb [57]. Sugar content is an essential component of fruit organoleptic quality, and thus is given a high priority in apple breeding programs. However, sugar content is inherited quantitatively and few genes responsible for fruit sugar accumulation have been identified although genetic mapping of quantitative trait loci (QTL) for fruit sugar content has been extensively conducted in apple [58,59,60,61,62]. To facilitate understanding of the complex mechanism controlling sugar accumulation in fruit, we investigated the roles of the SUT gene family in determining sugar content of apple fruit and an MdSUT4 gene was found to participate in regulation of fruit sugar accumulation. This study will assist us in understanding of the impact of SUT4 proteins on fruit sugar accumulation. The SUT4 gene along with its molecular tag can also serve as molecular tools that are useful for genetic improvement of fruit sweetness in breeding programs of apple.
Results
Identification of the SUT gene family in the apple genome
Screening the apple reference genome sequences of the ‘Golden Delicious’ doubled-haploid GDDH13 revealed a total of six MdSUT genes, which are located on different chromosomes (Fig. 1a). The deduced proteins of MdSUTs all contained 12 putative transmembrane α-helices (Fig. S1). Phylogenetic analysis showed that the six MdSUTs could be classified into three subfamilies, with each containing two members (Fig. 1b), and the MdSUT genes were subsequently named according to the standard gene nomenclature [1]. Interestingly, two MdSUT genes of the same subfamily shared similar genomic structure and were located on a pair of homologous chromosomes (Fig. 1a, c). The MdSUT1, MdSUT2, and MdSUT4 genes contained four, fourteen and five exons, and were located on three homologous pairs of chromosomes, 5–10, 13–16 and 8–15, respectively. In addition, the MdSUT2 proteins had a large central loop between TM6 and TM7, with a length of over 90 amino acids, whereas, the central loop length was less than 45 amino acids for both MdSUT1 and MdSUT4 (Fig. S1). Likewise, the length of N-terminal domain of MdSUT2 was ~ 2.7 times longer than those of MdSUT1 and MdSUT4. These results are consistent with the previous report that the SUT2 subfamily has the extended N-terminus and central loop [13].
Genes encoding sucrose transporter (SUT) in the apple genome. a Chromosomal location of the apple MdSUTs and their accession numbers in the Genome Database for Rosaceae (GDR). b Phylogenetic tree derived from amino acid sequences of SUTs in apple, peach and Arabidopsis. Numbers near branches indicate bootstrap test result with 1000 replicate analyses. C, Genomic structure of the MdSUT genes
Association between MdSUTs and sugar accumulation in apple fruit
Simple sequence repeat (SSR) markers were developed for MdSUT1.1 and MdSUT2.1 based on two intronic microsatellites, (CAAAAA)n and (TA)n, respectively (Table 1). Two microsatellites located upstream of the start codon, (CT)n and (AG)n, were successfully used to develop SSR markers for MdSUT1.2 and MdSUT4.1, respectively. Whereas, SSR markers for MdSUT4.2 and MdSUT2.2 were developed based on (TA)n and (CT)n microsatellites, respectively, located downstream of the stop codon. To estimate the association of the SUT genes with fruit sugar content, these newly developed SSR markers were used to genotype the 353 apple accessions reported in our previous study [63]. As a result, four polymorphic bands were detected for both MdSUT1.2 and MdSUT2.2, whereas, three, three, two and two polymorphic bands were identified for MdSUT1.1, MdSUT4.1, MdSUT4.2 and MdSUT2.1, respectively (Fig. S2a, b). Finally, association of the polymorphic loci of MdSUT genes with fruit sugar content was analyzed using the candidate gene-based association strategy. The results showed only the (AG)n microsatellite located 15 bp upstream of the start codon of MdSUT4.1 was significantly associated with fructose content, with the probability values (P-value) of 0.0028 (Table 1). Three alleles, (AG)6, (AG)9, and (AG)11, were identified at the MdSUT4.1 locus by direct sequencing of PCR products (Fig. 2a). Based on the present or absence of the (AG)9 on the MdSUT4.1 locus, all the accessions were divided into three genotypes: (AG)9/9, (AG)6/9, and (AG)6/6 or 11. Accessions with one or two (AG)9 alleles had significantly lower fructose content than those with no (AG)9 allele (Fig. 2b). However, the (AG)9 allele showed no significant impact on either total sugar or sucrose contents.
The (AG)n motif site (a) and the mean values of sugar contents in apple mature fruits of three different genotypes at the MdSUT4.1 locus among tested cultivars (b). Statistical significance was analyzed by Student’s t-test. **, P < 0.01; NS, no significant difference (P > 0.05). Error bars represent standard deviation of each genotype data set
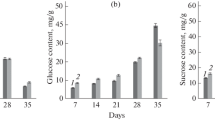
To further confirm the association of the MdSUT4.1 gene with apple fruit sugar accumulation, we investigated the expression profile of MdSUT4.1 in fruits of three cultivars and one crabapple at different stages of development (Fig. 3). Overall, the MdSUT4.1 gene was expressed most highly at the young stage, and then showed a decrease in expression throughout fruit development, with approximately 7- to 9-fold decrease at the mature stage. By contrast, sugar accumulation showed a positive trend throughout fruit development, with a peak at the mature stage (Fig. 3). In addition, the expression level of MdSUT4.1 was significantly negatively correlated with glucose content (r = − 0.359, P < 0.05), fructose content (r = − 0.618, P < 0.001), sucrose content (r = − 0.604, P < 0.001), and total sugar content (r = − 0.624, P < 0.001). Taken together, these results suggested that MdSUT4.1 was a candidate involved in fruit sugar accumulation and further subjected to functional analysis.
Expression profile of the MdSUT4.1 gene (a) and sugar accumulation (b) in fruits of three apple cultivars and one crabapple at various developmental stages. S1-S3 indicate young (60 DAF), expanding (90 DAF) and mature stages (120 DAF), respectively. Error bars represent standard error (SE) of three independent biological replicates
Subcellular localization of MdSUT4.1 in tobacco
As mentioned above, MdSUT4.1 contained twelve putative transmembrane domains. To further determine subcellular localization of MdSUT4.1, the yellow fluorescent protein (YFP) was fused to the C-terminus of MdSUT4.1. The MdSUT4.1-YFP fusion construct driven by CaMV 35S promoter was transiently expressed in tobacco (Nicotiana benthamiana), and the MdSUT4.1-YFP fusion protein was found to reside in the tonoplast (Fig. 4). In addition, we did co-localization with the standard vacuolar membrane marker Vac-rk CD3–975 to confirm the subcellular location of MdSUT4.1 (Fig. 4). Laser-scanning confocal fluorescence microscopy revealed that the mCherry fluorescence from the tonoplast marker Vac-rk CD3–975 was merged with the YPF fluorescence from MdSUT4.1-YFP. This result indicated that MdSUT4.1 is a tonoplast localized protein.
Overexpression of the MdSUT4.1 gene in strawberry and apple callus
To examine whether MdSUT4.1 was able to transport sugars, it was transiently transferred into immature white fruit of strawberry (Fig. 5a). Quantitative real time PCR (qRT-PCR) analysis showed that MdSUT4.1 was highly expressed in transgenic fruit (Fig. 5b). Sucrose content decreased significantly by 35% in fruit overexpressing MdSUT4.1 compared to fruit infiltrated with empty vector (Fig. 5c). However, the glucose, fructose, and total sugar contents showed no significant difference between fruits overexpressing MdSUT4.1 or introducing empty vector. qRT-PCR was conducted to investigate expression levels of genes related to sucrose metabolism or transport, including FaSPS (sucrose phosphate synthase), FaSUSY (sucrose synthase), FavAINV (vacuolar acid invertase) and FaSUT1, which are proven to play important roles in determining fruit sucrose content in strawberry [40, 64]. As a result, expression levels of FaSPS and FaSUSY were significantly lower in fruits overexpressing MdSUT4.1 than in fruits infiltrated with empty vector, but no significant difference in expression level was observed for FaSUT1 and FavINV (Fig. 6). This was consistent with the result that sucrose accumulation was decreased in fruits overexpressing MdSUT4.1.
Overexpression of MdSUT4.1 in strawberry fruit. a Strawberry fruits infiltrated individually with MdSUT4.1 (transgenic) and the entry vector pSAK277 (CK). Scale bar, 1 cm. b Relative expression of MdSUT4.1 in strawberry fruits infiltrated with MdSUT4.1 and the entry vector, respectively. c Sugar accumulation in strawberry fruits infiltrated with MdSUT4.1 and the entry vector, respectively. Statistical significance was analyzed by Student’s t-test. **, P < 0.01; NS, no significant difference (P > 0.05). Error bars represent SE of three biological replicates
Expression of sugar metabolism and sugar transporter genes in strawberry fruits infiltrated with MdSUT4.1 (black column) and the entry vector pSAK277 (control, gray column). Statistical significance of differences between mean values was analyzed by Student’s t-test. **, P < 0.01; *, P < 0.05; NS, no significant difference (P > 0.05). Error bars represent standard error (SE) of three biological replicates. SPS, sucrose phosphate synthase; SUSY, sucrose synthase; vAINV, vacuolar acid invertase
To further confirm the function of the MdSUT4.1 gene involved in fruit sugar accumulation, it was also transferred into the apple callus (Fig. 7a). qRT-PCR assay showed that the MdSUT4.1 gene was highly expressed in transgenic lines (Fig. 7b). The concentrations of sucrose in calli overexpressing MdSUT4.1 increased significantly by 1.77 mg/g FW, whilst the concentrations of glucose, fructose, and total sugars dramatically dropped by 27.96, 20.68, and 48.88 mg/g FW, respectively, compared to calli introducing the empty vector (Fig. 7c). qRT-PCR analysis was conducted to investigate expression levels of sugar transport-related genes (Fig. 8). As a result, a tonoplast sugar transporter MdTST1 and MdSUT2.2 showed significantly lower levels of expression in calli overexpressing MdSUT4.1 than in calli introducing the empty vector, but the opposite trend was observed for MdSUT4.2, MdSUT1.1 and MdSUT1.2. By contrast, MdSUT2.1 showed no significant difference in expression level between calli overexpressing MdSUT4.1 and calli introducing the empty vector. Moreover, expression levels of sugar metabolism-related genes that were reported to affect fruit sugar accumulation in apple [65] were also investigated (Fig. 9). MdvAINV1 showed significantly lower levels of expression in calli overexpressing MdSUT4.1 than in calli introducing the empty vector, while the opposite trend was observed for MdSPS and MdSDH2–9 (sorbitol dehydrogenase) genes. By contrast, MdNINV1 (Neutral/cytosol invertase), MdSUSY2, and MdCWINV2 (cell wall invertase) showed no significant difference in expression level between calli overexpressing MdSUT4.1 and calli introducing the empty vector.
Overexpression of MdSUT4.1 in apple calli. a Apple calli infiltrated with MdSUT4.1 (transgenic) and the entry vector pSAK277 (CK), respectively. b Expression of MdSUT4.1 in transgenic calli and CK. c Sugar accumulation in transgenic calli and CK. Statistical significance was analyzed by Student’s t-test. **, P < 0.01. Error bars represent SE of three biological replicates
Expression of MdTST1 and MdSUTs in apple calli infiltrated with MdSUT4.1 (black column) and the entry vector pSAK277 (control, gray column). Statistical significance was analyzed by Student’s t-test. Asterisk **, P < 0.01; *, P < 0.05; NS, no significant difference (P > 0.05). Error bars represent standard error (SE) of three biological replicates
Relative expression level of sugar metabolic gene in apple calli infiltrated with MdSUT4.1 (black column) and the entry vector pSAK277 (control, gray column). Statistical significance was analyzed by Student’s t-test. **, P < 0.01; *, P < 0.05; NS, no significant difference (P > 0.05). Error bars represent SE of three biological replicates. SPS, sucrose phosphate synthase; SDH, sorbitol dehydrogenase; vAINV, vacuolar acid invertase; NINV, neutral/cytosol invertase; CWINV, cell wall invertase; SUSY, sucrose synthase
Discussion
The complexity of mechanisms underlying the targeting of SUT4 proteins to the tonoplast
In fruit, soluble sugars are mainly stored in the cell vacuole, and their uptake into or release out of the vacuole is catalyzed by tonoplast-localized transporters. A di-leucine motif (LXXLL) within the N- and/or C-terminal domains is responsible for the targeting of sugar transporters to the tonoplast in plants [11, 66, 67]. A variety of SUT4 subfamily members that contain the di-leucine motif, such as Arabidopsis AtSUT4 (also known as AtSUC4) [24], Lotus japonicas LjSUT4 [25], barley HvSUT2 [24], rice OsSUT2 [68] and Populus PtaSUT4 [26], have been shown to reside in the vacuolar membrane. In fruit trees, PpSUT4 in peach, MdSUT4.1 and MdSUT4.2 in apple and PbSUT2 in pear all contain the vacuolar targeting di-leucine motif LXXLL (Fig. S3), thus, they are expected to localize in the tonoplast. MdSUT4.2 is also known as MdSUT1 in a previous report [33]. Like MdSUT4.1, PpSUT4 was shown to reside in the vacuolar membrane by its transient expression in tobacco leaf [29]. However, both MdSUT1 and PbSUT2 were shown to localize in the plasma membrane via their ectopic expression in Arabidopsis protoplasts and onion epidermis cells [33, 52]. This inconsistency indicates that other unknown factors, besides the di-leucine motif, may also influence subcellular localization of SUT4 subfamily members.
MdSUT4.1 participates in regulation of apple fruit sugar accumulation
Our study showed that MdSUT4.1 was significantly associated with fruit sugar accumulation in apple and is located on an interval of the sweetness QTL on LG8 [58]. The expression levels of MdSUT4.1 displayed a negative correlation with fruit sugar accumulation, and its overexpression in strawberry and apple callus showed an overall negative impact on sugar accumulation. These results demonstrate that the MdSUT4.1 gene is a strong candidate for the regulation of fruit sugar accumulation in apple.
As mentioned above, MdSUT4.1 resides in the vacuolar membrane, and its transient overexpression in strawberry significantly decreased sucrose accumulation in fruit. This suggests that MdSUT4.1 has similar functionality as Arabidopsis AtSUT4 and maize ZmSUT2, both of which are SUT4 subfamily members and function to remobilize sucrose out of the vacuole [27, 28, 69]. This hypothesis is consistent with the finding that the expression of sucrose synthesis gene FaSPS showed a significant decrease in strawberry fruit overexpressing MdSUT4.1 compared with fruit infiltrated with the empty vector. This change in expression level of FaSPS may be attributed to feedback from the efflux of sucrose from the vacuole caused by overexpression of MdSUT4.1.
Similarly, the overexpression of MdSUT4.1 in apple callus resulted in a significant decrease in hexose accumulation, which is consistent with the finding that the expression of MdSUT4.1 shows a negative correlation with sugar accumulation in apple fruit. MdSUT4.1 probably also mediates the efflux of sucrose from the vacuole in apple calli as this hypothesis is well consistent with the result that the expression of MdvAINV1 responsible for sucrose cleavage in the vacuole significantly decreased in apple calli overexpressing MdSUT4.1. The slight increase of sucrose accumulation in apple calli overexpressing MdSUT4.1 may be due to activation of MdSUT1.1 and MdSUT1.2 as the SUT1 genes are known to serve as the high-affinity sucrose transporters and function in sucrose uptake into sink storage tissues [70, 71].
Previous studies have demonstrated that SUT4 can interact with other sugar transporters such as SUT1 and TST [29, 43, 72]. Thus, it cannot be excluded that overexpression of MdSUT4.1 in apple callus may affect its interaction with other sugar transporter such as MdTST, resulting in the change in sugar accumulation. In addition, our results show that overexpression of MdSUT4.1 results in a cascade response of sugar metabolism and transporter genes to affect sugar accumulation, which is similar to previous reports [32, 73].
In apple, sorbitol is the predominant sugar in the phloem, but it only accounts for about 5% in mature fruit as most is converted to fructose [74]. The expression of genes encoding sorbitol transporter (SOT), including MdSOT1, MdSOT4 and MdSOT5.3, decreased in apple calli overexpressing MdSUT4.1 (Fig. S4). Thus, it is worthy of further study to ascertain whether overexpression of MdSUT4.1 has an impact on expression of MdSOTs, resulting in a change in sugar accumulation in apple fruit.
Conclusions
Collectively, our results show that the tonoplast-localized sucrose transporter, MdSUT4.1, participates in regulating fruit sugar accumulation in apple. Our results are useful for better understanding of the impact of SUT4 proteins on fruit sugar accumulation. In addition, the molecular marker of MdSUT4.1 may serve as a tool for genetic improvement of fruit sweetness in breeding programs of apple.
Methods
Plant material
All apple accessions used in this study are maintained at Xingcheng Institute of Pomology, the Chinese Academy of Agricultural Sciences, Xingcheng, Liaoning province, China. Young leaves were collected for each cultivar, frozen in liquid nitrogen, and then stored at − 80 °C until use. Fruit samples for sugar content measurement were randomly harvested at mature stage in 2015, which was estimated based on previous records of maturity date, skin background color and blush development, and seed color changing to brown. Moreover, one crab apple (Malus coronaria) and three cultivars, ‘Calville Rouge’, ‘Lisijin’, and ‘Weiqinni’, were selected for qRT-PCR analysis. Fruit samples were harvested at young, immature and mature stages, which corresponded to 60, 90, and 120 days after flowering (DAF), respectively. Three biological replicates with 10 fruits were conducted for each accession. Fruit samples were frozen in liquid nitrogen and stored either at − 80 °C for qRT-PCR analysis or at − 40 °C for sugar measurement. In addition, strawberry cultivar ‘Hongyan’ was kindly provided by Institute of Economic Crops, Hubei Academy of Agricultural Sciences, Wuhan, China and it was grown in the greenhouse with natural light and temperature of 10 °C – 26 °C. Apple calli induced from flesh of apple cultivar ‘Orin’ was kindly provided by Prof. Qingmei Guan at Northwest Agriculture University, Yangling, China.
Measurement of sugar contents in fruits
Sugar content was measured by high performance liquid chromatography (HPLC) according to our previously reported protocol [75]. Total sugars in this study represents the sum of dominant soluble sugars, including sucrose, fructose and glucose, which together account for over 95% of the total sugar in apple fruits [74]. Briefly, fruit samples were ground into powder in liquid nitrogen using an A11 Basic Analytical Mill (IKA-Werke, Staufen, Germany) and approximately 0.5 g of powder was transferred to a clean Eppendorf tube. Soluble sugars were extracted with 6 mL deionized water by ultrasonic treatment for 15 min, and then centrifuged at 5000×g for 15 min. The resultant supernatant was filtered through a 0.22 μm Sep-Pak filter (ANPEL, China). The filters were subjected to sugar content measurement using HPLC system (Agilent 1260 Infinity, Agilent, Waldbronn, Germany) with refractive index detector (RID). The injection volume was 20 μL, and chromatographic separation was performed using a CarboSep CHO-620 Ca column (300 × 6.5 mm, 10 μm particle size) and CarboSep CHO-620 Cartridge (Transgenomic, San Jose, CA, USA). The mobile phase was deionized water at the flow rate of 0.5 mL/min and the column temperature was maintained at 90 °C. Sugar concentration was estimated by comparison with the values from a standard curve. Sucrose (CAS number 57–50-1), glucose (CAS number 50–99-7), and fructose (CAS number 57–48-7) standards were purchased from Sigma. Each accession had three biological replicates.
RNA isolation and qRT-PCR analysis
Total RNA was extracted with the RNAprep pure Plant kit (Tiangen, Beijing, China) following the manufacturer’s instructions, and genomic DNA contamination was removed using DNase I (Takara, Dalian, China). The synthesis of cDNA templates was conducted using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TRANS, Beijing, China) following the manufacturer’s instructions. The qRT-PCR assay was carried out with TB Green® Premix Ex Taq™ II (Tli RNase H Plus) (Takara, Dalian, China), and amplifications were performed on Applied Biosystems Step One Plus Real-Time PCR Systems (Life Technologies Corporation, Carlsbad, CA, USA). Melting curve was analyzed at the end of 40 cycles to confirm specific amplification products. A previously reported actin gene [76] was used as a constitutive control, and negative control for each sample was also performed. Each sample had three biological replicates. Primer sequences are listed in Table S1.
Identification of SUT genes in the apple genome and their classification
Identification of SUT genes was initially conducted based on the apple genome annotation database for GDDH13 [77], and six MdSUT genes were identified. Subsequently, the coding sequence of each SUT gene was further compared against the apple draft genome using the online program BlastN (https://www.rosaceae.org/blast), with an E value cutoff of 1e-10, and no additional member was identified. To validate the annotation of the MdSUT genes, their protein sequences were BLASTed against GenBank’s non-redundant protein (NR) and Swiss-Prot protein databases using BLASTp, an E value cutoff of 1e-5. Chromosomal locations of MdSUTs were presented according to the draft genome sequence of GDDH13 [77]. Prediction of protein transmembrane domains was conducted using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) [78]. Gene structures were drawn using the visualized software IBS Version 1.0 [79].
To classify MdSUTs, their amino-acid sequences were aligned with those of SUT genes from Arabidopsis and peach using MEGA5 with default parameters. The resulting data matrix was used to construct phylogenetic tree using the Neighbor-Joining method. Bootstrap values were estimated from 1000 replicates.
Development of gene-tagged markers and candidate gene-based association analysis
Genomic DNA sequences of each MdSUT were retrieved from the draft genome of GDDH13 [77] and then subjected to screen SSRs with ≥6 repeat units using microsatellite repeats finder (http://insilico.ehu.eus/mini_tools/microsatellites/). Primers of SSR markers were designed using Primer premier 5. SSR polymorphism was detected using polyacrylamide gel electrophoresis in combination with silver staining following a previous report [80].
Polymorphic SSRs for MdSUTs were used to screen the same collection of 353 apple accessions as in our previous study [63], and their primer sequences are listed in Table S2. Association of MdSUTs with fruit sugar concentration was carried out with TASSEL version 3.0 using a mixed linear model, with P < 0.01 as the criterion for significant marker–trait association. The relative kinship (K matrix) and the most likely K value (Q matrix) were the same in our previous study [63]. The Bonferroni threshold was set at ≤1/n, where n represents the sample size.
Subcellular localization analysis of MdSUT4.1 in tobacco
YFP fragment was amplified from pEarleyGate104 and then cloned into the BamHI-SpeI sites of pFGC5941 using Hieff CloneTM Plus One Step Cloning Kit (Yeasen, Shanghai, China), resulting in a pFGC-eYFP vector. Amplification of the full-length coding sequences of MdSUT4.1 was performed using cDNA templates that were prepared from fruits of ‘Calville Rouge’. The purified PCR products were inserted into the BamHI site of the pFGC-eYFP vector to generate a pFGC-eYFP-MdSUT4.1 construct using In-Fusion® HD Cloning Kit (Takara, Dalian, China) following the manufacturer’s instructions. Primer sequences used for expression vector construction are listed in Table S3.
The vacuolar membrane marker vac-rk CD3–975 [81] and the pFGC-eYFP-MdSUT4 construct were individually transferred into Agrobacterium tumefaciens strain GV3101 (pMP90) using the heat shock method, spread onto agar plates, and incubated at 28 °C for 3 days. A single colony was picked, suspended in LB medium, and incubated at 28 °C overnight under shaking. Agrobacterium cells were collected by centrifugation at 4000 g for 5 min and resuspended in infiltration buffer (10 mM MES, 10 mM MgCl2, 450 μM acetosyringone, pH 5.6). Leaves of 4–5 weeks old N. benthamiana were infiltrated with bacterial cultures and the fluorescence was detected two days after infiltration using the confocal microscope (TCS SP8, Leica, Microsystems, Wetzlar, Germany). All scanners used a × 20 objective lens to acquire the digital images. A 512 nm laser was used for the excitation of YFP, while mCherry was excited by 552 nm laser. Fluorescence emission was detected in the ranges of 530–550 nm (YFP) and 600–650 nm (mCherry), respectively.
Functional analysis of MdSUT4.1 in strawberry and transgenic apple callus
The entire CDS of MdSUT4.1 was amplified and inserted into the site of the expression vector pSAK277 between EcoRI and XbaI enzyme sites via homologous recombination. Primer sequences are listed in Table S3. The recombinant vector pSAK277-MdSUT4.1 and the empty vector pSAK277 were individually transferred into A. tumefaciens strain EHA105 with the heat shock method, and then spread onto agar plates containing 50 mg/L streptomycin, 50 mg/L gentamycin and 100 mg/L spectinomycin. After incubation at 28 °C for 3 days, single colony was picked, suspended in LB medium, and incubated at 28 °C overnight with shaking. Agrobacterium cells were collected by centrifugation at 4000 g for 5 min and then used to infiltrate fruit of strawberry cv. Hongyan and apple calli that were induced from flesh of cv. Orin.
Transient overexpression of MdSUT4.1 in strawberry was conducted according to a previous report [82]. Briefly, the collected bacterial pellets were resuspended in infiltration buffer (50 mM MES, 5 mg/mL D-glucose, 2 mM Na3PO4, 100 μM acetosyringone) and the density of cells was adjusted to an OD 600nm of 0.2. The suspension was injected into the flesh of immature white fruits with needle of syringes, and the infiltrated plants were placed in the greenhouse with natural light and temperature of 10 °C–26 °C. Fruit samples were collected 9 days after infiltration and then subjected to qRT-PCR analysis and sugar content measurement.
For apple callus transformation, the collected bacterial pellets were resuspended in 5% sucrose solution (pH 5.3) and the density of cells was adjusted to an OD 600nm of 2.0. Apple calli were immerged in the suspension for 15 min, collected by filtration through sterile nylon net (200 mesh), and then cultured on MS medium containing 3% sucrose, 1 mg/L 2,4-D, 1 mg/L 6-BA and 400 mg/L cefalexin for two days at 25 °C under dark condition. Subsequently, the transgenic apple calli were washed 5 times with sterile Mili-Q water containing 400 mg/L cefalexin, dried on filter, and then cultured on MS selection medium containing 3% sucrose, 1 mg/L 2,4-D, 1 mg/L 6-BA, 400 mg/L cefalexin and 50 mg/L Kanamycin. After successive subculture for three callus generations, transgenic calli were subjected to sugar content measurement using HPLC. In addition, transgenic calli were confirmed by PCR amplification and expression of MdSUT4.1 was quantified using qRT-PCR analysis. Primer sequences are listed in Table S1.
Availability of data and materials
The datasets used and analyzed during the current study available from the corresponding author on reasonable request.
Abbreviations
- SUT:
-
Sucrose transporter
- TST:
-
Tonoplast sugar transporter
- SOT:
-
Sorbitol transporter
- SPS:
-
Sucrose phosphate synthase
- SDH:
-
Sorbitol dehydrogenase
- vAINV:
-
Vacuolar acid invertase
- NINV:
-
Neutral/cytosol invertase
- CWINV:
-
Cell wall invertase
- SUSY:
-
Sucrose synthase
- DAF:
-
Days after flowering
- qRT-PCR:
-
Quantitative real time PCR
- HPLC:
-
High performance liquid chromatography
References
Kühn C, Grof CPL. Sucrose transporters of higher plants. Curr Opin Plant Biol. 2010;13:288–98.
Sauer N. Molecular physiology of higher plant sucrose transporters. FEBS Lett. 2007;581:2309–17.
Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH Jr. The major facilitator superfamily (MFS) revisited. FEBS J. 2012;279:2022–35.
Yan N. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem Sci. 2013;38:151–9.
Chandran D, Reinders A, Ward JM. Substrate specificity of the Arabidopsis thaliana sucrose transporter AtSUC2. J Biol Chem. 2003;278:44320–5.
Carpaneto A, Geiger D, Bamberg E, Sauer N, Fromm J, Hedrich R. Phloem-localized, proton-coupled sucrose carrier ZmSUT1 mediates sucrose efflux under the control of the sucrose gradient and the proton motive force. J Biol Chem. 2005;280:21437–43.
Riesmeier JW, Willmitzer L, Frommer WB. Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 1992;11:4705–13.
Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815.
Aoki N, Hirose T, Scofield GN, Whitfeld PR, Furbank RT. The sucrose transporter gene family in Rice. Plant Cell Physiol. 2003;44:223–32.
Lalonde S, Frommer WB. SUT sucrose and MST monosaccharide transporter inventory of the Selaginella genome. Front Plant Sci. 2012;3:24.
Reinders A, Sivitz AB, Ward JM. Evolution of plant sucrose uptake transporters (SUTs). Front Plant Sci. 2012;3:22.
Peng D, Gu X, Xue LJ, Leebens-Mack JH, Tsai CJ. Bayesian phylogeny of sucrose transporters: ancient origins, differential expansion and convergent evolution in monocots and dicots. Front Plant Sci. 2014;5:615.
Barker L, Kühn C, Weise A, Schulz A, Gebhardt C, Hirner B, Hellmann H, Schulze W, Ward JM, Frommer WB. SUT2, a putative sucrose sensor in sieve elements. Plant Cell. 2000;12:1153–64.
Riesmeier JW, Willmitzer L, Frommer WB. Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J. 1994;13:1–7.
Bürkle L, Hibberd JM, Quick WP, Kühn C, Hirner B, Frommer WB. The H+-sucrose cotransporter NtSUT1 is essential for sugar export from tobacco leaves. Plant Physiol. 1998;118:59–68.
Shakya R, Sturm A. Characterization of source- and sink-specific sucrose/H+ symporters from carrot. Plant Physiol. 1998;118:1473–80.
Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR. Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc Natl Acad Sci U S A. 2000;97:13979–84.
Öner-Sieben S, Lohaus G. Apoplastic and symplastic phloem loading in Quercus robur and Fraxinus excelsior. J Exp Bot. 2014;65:1905–16.
Öner-Sieben S, Rappl C, Sauer N, Stadler R, Lohaus G. Characterization, localization, and seasonal changes of the sucrose transporter FeSUT1 in the phloem of Fraxinus excelsior. J Exp Bot. 2015;66:4807–19.
Nieberl P, Ehrl C, Pommerrenig B, Graus D, Marten I, Jung B, Ludewig F, Koch W, Harms K, Flugge UI, Neuhaus HE, Hedrich R, Sauer N. Functional characterisation and cell specificity of BvSUT1, the transporter that loads sucrose into the phloem of sugar beet (Beta vulgaris L.) source leaves. Plant Biol. 2017;19:315–26.
Dobbelstein E, Fink D, Öner-Sieben S, Czempik L, Lohaus G. Seasonal changes of sucrose transporter expression and sugar partitioning in common European tree species. Tree Physiol. 2018;39:284–99.
Ma QJ, Sun MH, Lu J, Liu YJ, Hu DG, Hao YJ. Transcription factor AREB2 is involved in soluble sugar accumulation by activating sugar transporter and amylase genes. Plant Physiol. 2017;174:2348–62.
Ma QJ, Sun MH, Lu J, Kang H, You CX, Hao YJ. An apple sucrose transporter MdSUT2.2 is a phosphorylation target for protein kinase MdCIPK22 in response to drought. Plant Biotechnol J. 2019;17:625–37.
Endler A, Meyer S, Schelbert S, Schneider T, Weschke W, Peters SW, Keller F, Baginsky S, Martinoia E, Schmidt UG. Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol. 2006;141:196–207.
Reinders A, Sivitz AB, Starker CG, Gantt JS, Ward JM. Functional analysis of LjSUT4, a vacuolar sucrose transporter from Lotus japonicus. Plant Mol Biol. 2008;68:289–99.
Payyavula RS, Tay KHC, Tsai CJ, Harding SA. The sucrose transporter family in Populus: the importance of a tonoplast PtaSUT4 to biomass and carbon partitioning. Plant J. 2011;65:757–70.
Schulz A, Beyhl D, Marten I, Wormit A, Neuhaus E, Poschet G, Büttner M, Schneider S, Sauer N, Hedrich R. Proton-driven sucrose symport and antiport are provided by the vacuolar transporters SUC4 and TMT1/2. Plant J. 2011;68:129–36.
Schneider S, Hulpke S, Schulz A, Yaron I, Höll J, Imlau A, Schmitt B, Batz S, Wolf S, Hedrich R, Sauer N. Vacuoles release sucrose via tonoplast-localised SUC4-type transporters. Plant Biol. 2012;14:325–36.
Zanon L, Falchi R, Hackel A, Kühn C, Vizzotto G. Expression of peach sucrose transporters in heterologous systems points out their different physiological role. Plant Sci. 2015;238:262–72.
Weise A, Barker L, Kühn C, Lalonde S, Buschmann H, Frommer WB, Ward JM. A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. Plant Cell. 2000;12:1345–56.
Weschke W, Panitz R, Sauer N, Wang Q, Neubohn B, Weber H, Wobus U. Sucrose transport into barley seeds: molecular characterization of two transporters and implications for seed development and starch accumulation. Plant J. 2000;21:455–67.
Chincinska IA, Gier K, Krügel U, Liesche J, He HX, Grimm B, Harren FJM, Cristescu SM, Kühn C. Photoperiodic regulation of the sucrose transporter StSUT4 affects the expression of circadian-regulated genes and ethylene production. Front Plant Sci. 2013;4:26.
Fan RC, Peng CC, Xu YH, Wang XF, Li Y, Shang Y, Du SY, Zhao R, Zhang XY, Zhang LY, Zhang DP. Apple sucrose transporter SUT1 and sorbitol transporter SOT6 interact with cytochrome b5 to regulate their affinity for substrate sugars. Plant Physiol. 2009;150:1880–901.
Slewinski TL, Meeley R, Braun DM. Sucrose transporter1 functions in phloem loading in maize leaves. J Exp Bot. 2009;60:881–92.
Sun Y, Reinders A, LaFleur KR, Mori T, Ward JM. Transport activity of rice sucrose transporters OsSUT1 and OsSUT5. Plant Cell Physiol. 2010;51:114–22.
Lemoine R, Bürkle L, Barker L, Sakr S, Kühn C, Regnacq M, Gaillard C, Delrot S, Frommer WB. Identification of a pollen-specific sucrose transporter-like protein NtSUT3 from tobacco. FEBS Lett. 1999;454:325–30.
Stadler R, Truernit E, Gahrtz M, Sauer N. The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. Plant J. 1999;19:269–78.
Sivitz AB, Reinders A, Johnson ME, Krentz AD, Grof CPL, Perroux JM, Ward JM. Arabidopsis sucrose transporter AtSUC9. High-affinity transport activity, intragenic control of expression, and early flowering mutant phenotype. Plant Physiol. 2007;143:188–98.
Hackel A, Schauer N, Carrari F, Fernie AR, Grimm B, Kühn C. Sucrose transporter LeSUT1 and LeSUT2 inhibition affects tomato fruit development in different ways. Plant J. 2006;45:180–92.
Jia HF, Wang YH, Sun MZ, Li BB, Han Y, Zhao YX, Li XL, Ding N, Li C, Ji WL, Jia WS. Sucrose functions as a signal involved in the regulation of strawberry fruit development and ripening. New Phytol. 2013;198:453–65.
Sivitz AB, Reinders A, Ward JM. Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiol. 2008;147:92–100.
Kühn C, Hajirezaei MR, Fernie AR, Roessner-Tunali U, Czechowski T, Hirner B, Frommer WB. The sucrose transporter StSUT1 localizes to sieve elements in potato tuber phloem and influences tuber physiology and development. Plant Physiol. 2003;131:102–13.
Reinders A, Schulze W, Kühn C, Barker L, Schulz A, Ward JM, Frommer WB. Protein–protein interactions between sucrose transporters of different affinities colocalized in the same enucleate sieve element. Plant Cell. 2002;14:1567–77.
Ahmed SAS, Zhang JJ, Ma WJ, Dell B. Contributions of TaSUTs to grain weight in wheat under drought. Plant Mol Biol. 2018;98:333–47.
Wang W, Zhou H, Ma BQ, Owiti A, Korban SS, Han YP. Divergent evolutionary pattern of sugar transporter genes is associated with the difference in sugar accumulation between grasses and eudicots. Sci Rep. 2016;6:29153.
Li CL, Meng D, Pineros MA, Mao YX, Dandekar AM, Cheng LL. A sugar transporter takes up both hexose and sucrose for sorbitol-modulated in vitro pollen tube growth in apple. Plant Cell. 2019;32:449–69.
Ageorges A, Issaly N, Picaud S, Delrotb S, Romieu C. Identification and functional expression in yeast of a grape berry sucrose carrier. Plant Physiol Biochem. 2000;38:177–85.
Zhang YL, Meng QY, Zhu HL, Guo Y, Gao HY, Luo YB, Lu J. Functional characterization of a LAHC sucrose transporter isolated from grape berries in yeast. Plant Growth Regul. 2007;54:71–9.
Cai YM, Tu WR, Zu YY, Yan J, Xu ZB, Lu J, Zhang YL. Overexpression of a grapevine sucrose transporter (VvSUC27) in tobacco improves plant growth rate in the presence of sucrose in vitro. Frontiers Plant Sci. 2017;8:1069.
Zheng QM, Tang Z, Xu Q, Deng XX. Isolation, phylogenetic relationship and expression profiling of sugar transporter genes in sweet orange (Citrus sinensis). Plant Cell Tiss Org. 2014;119:609–24.
Zanon L, Falchi R, Santi S, Vizzotto G. Sucrose transport and phloem unloading in peach fruit: potential role of two transporters localized in different cell types. Physiol Plantarum. 2015;154:179–93.
Wang LF, Qi XX, Huang XS, Xu LL, Jin C, Wu J, Zhang SL. Overexpression of sucrose transporter gene PbSUT2 from Pyrus bretschneideri, enhances sucrose content in Solanum lycopersicum fruit. Plant Physiol Bioch. 2016;105:150–61.
Peng CC, Xu YH, Xi RC, Zhao XL. Expression, subcellular localization and phytohormone stimulation of a functional sucrose transporter (MdSUT1) in apple fruit. Sci Hortic. 2011;128:206–12.
Ma QJ, Sun MH, Liu YJ, Lu J, Hu DG, Hao YJ. Molecular cloning and functional characterization of the apple sucrose transporter gene MdSUT2. Plant Physiol Bioch. 2016;109:442–51.
Afoufa-Bastien D, Medici A, Jeauffre J, Coutos-Thévenot P, Lemoine R, Atanassova R, Laloi M. The Vitis vinifera sugar transporter gene family: phylogenetic overview and macroarray expression profiling. BMC Plant Biol. 2010;10:245.
Zhang CM, Bian Y, Hou SH, Li XG. Sugar transport played a more important role than sugar biosynthesis in fruit sugar accumulation during Chinese jujube domestication. Planta. 2018;248:1187–99.
Velasco R, Zharkikh A, Affourtit J, et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet. 2010;42:833–ss.
Liebhard R, Kellerhals M, Pfammatter W, Jertmini M, Gessler C. Mapping quantitative physiological traits in apple (Malus x domestica Borkh.). Plant Mol Biol. 2003;52:511–26.
Kenis K, Keulemans J, Davey MW. Identification and stability of QTLs for fruit quality traits in apple. Tree Genet Genomes. 2008;4:647–61.
Kunihisa M, Moriya S, Abe K, Okada K, Haji T, Hayashi T, Kim H, Nishitani C, Terakami S, Yamamoto T. Identification of QTLs for fruit quality traits in Japanese apples: QTLs for early ripening are tightly related to preharvest fruit drop. Breeding Sci. 2014;64:240–51.
Guan YZ, Peace C, Rudell D, Verma S, Evans K. QTLs detected for individual sugars and soluble solids content in apple. Mol Breeding. 2015;35:135.
Ma BQ, Zhao S, Wu BH, Wang DM, Peng Q, Owiti A, Fang T, Liao L, Ogutu C, Korban SS, Li SH, Han YP. Construction of a high density linkage map and its application in the identification of QTLs for soluble sugar and organic acid components in apple. Tree Genet Genomes. 2016;12:1.
Ma BQ, Liao L, Zheng HY, Chen J, Wu BH, Ogutu C, Li S, Korban SS, Han YP. Genes encoding aluminum-activated malate transporter II and their association with fruit acidity in apple. Plant Genome. 2015;8:1–14.
Tian L, Jia HF, Li CL, Fan PG, Xing Y, Shen YY. Sucrose accumulation during grape berry and strawberry fruit ripening is controlled predominantly by sucrose synthase activity. J Hortic Sci Biotech. 2012;87:661–7.
Li MJ, Feng FJ, Cheng LL. Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development. PLoS One. 2012;7:e33055.
Yamada K, Osakabe Y, Mizoi J, Nakashima K, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis thaliana abiotic stress-inducible facilitated diffusion transporter for monosaccharides. J Biol Chem. 2010;285:1138–46.
Wolfenstetter S, Wirsching P, Dotzauer D, Schneider S, Sauer N. Routes to the tonoplast: the sorting of tonoplast transporters in Arabidopsis mesophyll protoplasts. Plant Cell. 2012;24:215–32.
Eom JS, Cho JI, Reinders A, Lee SW, Yoo Y, Tuan PQ, Choi SB, Bang G, Park YI, Cho MH, Bhoo SH, An G, Hahn TR, Ward JM, Jeon JS. Impaired function of the tonoplast-localized sucrose transporter in rice, OsSUT2, limits the transport of vacuolar reserve sucrose and affects plant growth. Plant Physiol. 2011;157:109–19.
Leach KA, Tran TM, Slewinski TL, Meeley RB, Braun DM. Sucrose transporter2 contributes to maize growth, development, and crop yield. J Integr Plant Biol. 2017;59:390–408.
Rosche E, Blackmore D, Tegeder M, Richardson T, Schroeder H, Higgins TJ, Frommer WB, Offler CE, Patrick JW. Seed-specific overexpression of a potato sucrose transporter increases sucrose uptake and growth rates of developing pea cotyledons. Plant J. 2002;30:165–75.
Leggewie G, Kolbe A, Lemoine R, Roessner U, Lytovchenko A, Zuther E, Kehr J, Frommer WB, Riesmeier JW, Willmitzer L, Fernie AR. Overexpression of the sucrose transporter SoSUT1 in potato results in alterations in leaf carbon partitioning and in tuber metabolism but has little impact on tuber morphology. Planta. 2003;217:158–67.
Schulze WX, Reinders A, Ward J, Lalonde S, Frommer WB. Interactions between co-expressed Arabidopsis sucrose transporters in the split-ubiquitin system. BMC Biochem. 2003;4:3.
Chincinska IA, Liesche J, Krügel U, Michalska J, Geigenberger P, Grimm B, Kühn C. Sucrose transporter StSUT4 from potato affects flowering, tuberization, and shade avoidance response. Plant Physiol. 2008;146:515–28.
Yamaki S, Ino M. Alteration of cellular compartmentation and membrane permeability to sugars in immature and mature apple fruit. J Am Soc Hortic Sci. 1992;117:951–4.
Ma BQ, Chen J, Zheng HY, Fang T, Ogutu C, Li SH, Han YP, Wu BH. Comparative assessment of sugar and malic acid composition in cultivated and wild apples. Food Chem. 2015;172:86–91.
Ma BQ, Liao L, Fang T, Peng Q, Ogutu C, Zhou H, Ma FW, Han YP. A Ma10 gene encoding P-type ATPase is involved in fruit organic acid accumulation in apple. Plant Biotechnol J. 2019;17:674–86.
Daccord N, Celton JM, Linsmith G, et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet. 2017;49:1099–106.
Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202.
Liu WZ, Xie YB, Ma JY, Luo XT, Nie P, Zuo ZX, Lahrman U, Zhao Q, Zheng YY, Zhao Y, Xue X, Ren J. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics. 2015;31:3359–61.
Zhen QL, Fang T, Peng Q, Liao L, Zhao L, Owiti A, Han YP. Developing gene-tagged molecular markers for evaluation of genetic association of apple SWEET genes with fruit sugar accumulation. Hortic Res. 2018;5:14.
Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–36.
Cheng JT, Wen SY, Xiao S, Lu BY, Ma MR, Bie ZL. Overexpression of the tonoplast sugar transporter CmTST2 in melon fruit increases sugar accumulation. J Exp Bot. 2018;69:511–23.
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the National Key R&D Program of China (2018YFD1000200), the Special Fund for Strategic Pilot Technology of Chinese Academy of Sciences (XDA24030404–4), and National Natural Science Foundation of China (Grant No. 31420103914).
Author information
Authors and Affiliations
Contributions
QP conducted molecular marker development, gene expression and transgenic analysis, and prepared the manuscript. LL and QP did statistical analyses. YC, EL, and MN participated in gene functional studies. CO, SC, BZ, and YH revised the manuscript. YH was overall project leader. All authors read and approved the final manuscript.
Authors’ information
CAS Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, The Innovative Academy of Seed Design, Chinese Academy of Sciences, Wuhan, 430074 China.
Qian Peng, Yaming Cai, Enhui Lai, Liao Liao, Beibei Zheng, Collins Ogutu, Sylvia Cherono & Yuepeng Han.
University of Chinese Academy of Sciences, 19A Yuquanlu, Beijing 100049, China.
Qian Peng, Yaming Cai, Enhui Lai, Collins Ogutu & Sylvia Cherono.
Institute of Transformative Bio-Molecules, Nagoya University, Nagoya, Japan.
Masayoshi Nakamura.
Center of Economic Botany, Core Botanical Gardens, Chinese Academy of Sciences, Wuhan 430074, China.
Liao Liao, Beibei Zheng & Yuepeng Han.
Sino-African Joint Research Center, Chinese Academy of Sciences, Wuhan, 430074, China.
Yuepeng Han.
Corresponding author.
Correspondence to Yuepeng Han. Email: yphan@wbgcas.cn.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Sequences of primers used for qRT-PCR analysis.
Additional file 2: Table S2.
Primer sequences of gene-tagged SSR markers for six MdSUT genes in apple.
Additional file 3: Table S3.
Sequences of primers used for vector construction.
Additional file 4: Figure S1.
A schematic of the predicted topology of MdSUTs in apple. AA, amino acid.
Additional file 5:Figure S2.
An example of genotyping of apple cultivars using SSR markers of the MdSUT genes.
Additional file 6:
Figure S3. Alignment of vacuolar targeting di-leucine motif (LXXLL) in the N-terminus of SUT4 subfamily members in apple, peach, pear and Arabidopsis.
Additional file 7:
Figure S4. Expression of MdSOTs in apple calli overexpressing MdSUT4.1 (black column) and introducing entry vector pSAK277 (control, gray column).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Peng, Q., Cai, Y., Lai, E. et al. The sucrose transporter MdSUT4.1 participates in the regulation of fruit sugar accumulation in apple. BMC Plant Biol 20, 191 (2020). https://doi.org/10.1186/s12870-020-02406-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-020-02406-3