Abstract
Background
Avian pathogenic Escherichia coli (APEC) is the infectious agent of a wide variety of avian diseases, which causes substantial economic losses to the poultry industry worldwide. Polyamines contribute to the optimal synthesis of nucleic acids and proteins in bacteria. The objectives of this study were to investigate; i) whether APEC E. coli encodes the same systems for biosynthesis and uptake as described for E. coli K12 and ii) the role of polyamines during in vitro growth of an avian pathogenic E. coli strain (WT-ST117- O83:H4T).
Results
Following whole genome sequencing, polyamine biosynthesis and export genes present in E. coli MG1655 (K-12) were found to be identical in WT-ST117. Defined mutants were constructed in putrescine and spermidine biosynthesis pathways (ΔspeB, ΔspeC, ΔspeF, ΔspeB/C and ΔspeD/E), and in polyamines transport systems (ΔpotE, ΔyeeF, ΔpotABCD and ΔpotFGHI). Contrary to what was observed for MG1655, the ΔpotE-ST117 mutant was growth attenuated, regardless of putrescine supplementation. The addition of spermidine or orthinine restored the growth to the level of WT-ST117. Growth attenuation after induction of membrane stress by SDS suggested that PotE is involved in protection against this stress. The ΔspeB/C-ST117 mutant was also growth attenuated in minimal medium. The addition of putrescine or spermidine to the media restored growth rate to the wild type level. The remaining biosynthesis and transport mutants showed a growth similar to that of WT-ST117. Analysis by Ultra-High Performance Liquid Chromatography revealed that the ΔspeB/C mutant was putrescine-deficient, despite that the gene speF, which is also involved in the synthesis of putrescine, was expressed.
Conclusions
Deletion of the putrescine transport system, PotE, or the putrescine biosynthesis pathway genes speB/C affected in vitro growth of APEC (ST117- O83:H4) strain, but not E. coli MG1655, despite the high similarity of the genetic make-up of biosynthesis and transport genes. Therefore, blocking these metabolic reactions may be a suitable way to prevent APEC growth in the host without disturbing the commensal E. coli population.
Similar content being viewed by others
Background
Avian pathogenic (APEC) Escherichia coli strains belong to the group of extraintestinal pathogenic E. coli (ExPEC) [1]. They are the cause of avian colibacillosis [2], which is characterized by multiple organ lesions as airsacculitis, polyserositis, and pericarditis [3]. In recent years, a particular APEC strain has been associated with severe disease outbreaks in the Danish broiler production [4]. This strain, E. coli 083:H4 is assigned to the sequence type ST117 and the phylogenetic group D [5].
Polyamines, including putrescine, spermidine, spermine, and cadaverine, are cationic molecules derived from amino acids. Polyamines play an important role in the cell metabolism [6]. They can be involved in bacterial cell-cell signalling [7], motility [8], and are necessary for proper cell division [9]. They also contribute to the optimal synthesis of nucleic acids and proteins used in different growth processes [10]. Studies performed in Salmonella [11, 12], Legionella [13], Francisella [14] and Shigella [15] have demonstrated that polyamines are essential for virulence of these intracellular pathogens. Due to this variety of effects, polyamine metabolic pathways may represent a potential novel target for antibiotics [16, 17].
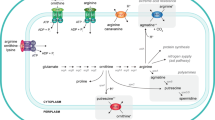
Multiple putrescine uptakes and degradation systems have been described in E. coli K-12, although some associated functions still remain to be completely understood. Intracellular polyamines amounts are regulated by biosynthesis, degradation, excretion, and uptake from the surrounding environment [18]. Putrescine is synthesized from L-arginine by the reactions which are catalysed by the enzymes arginine decarboxylase and agmatinase enzymes. These enzymes are encoded by the genes speA and speB. Putrescine can also be synthesized from L-ornithine through the action of the two ornithine-decarboxylation enzymes (OCDs), which are encoded by either speC or speF (Fig. 1) [19, 20]. The substrates for spermidine biosynthesis are putrescine and decarboxylated S-adenosyl-methionine (SAM) and requires enzymes encoded by speD and speE. Regulation of these biosynthesis processes is not fully understood; however, the intracellular amount of spermidine seems to be self-regulated [21].
Graphic representation of putrescine/spermidine biosynthesis pathways and transport systems a) putrescine/spermidine biosynthesis, where the deleted genes in the present study are indicated by an asterisk; b) putrescine/spermidine main transport systems, where ‘p’ represents putrescine and ‘s’ represents spermidine c) graphic representation of putrescine/proton symporter and putrescine/ornithine antiporter, where ‘p’ represents putrescine and ‘o’ represents ornithine. The figure is revised from Shah (20) and BioCyc (https://biocyc.org); SAM: S-adenosylmethionine; Decarbox. SAM: Decarboxylated SAM
The two major recognized systems for polyamine uptake in E. coli K12 belong to the family of ATP-binding-cassette (ABC) transporters. They are classified as the spermidine-preferential uptake system (PotABCD) and the putrescine-specific uptake system (PotFGHI) [21]. PotE is a membrane protein involved in the uptake of putrescine via proton symport mechanism, and it has been found to catalyze putrescine efflux by putrescine-ornithine antiport activity in E. coli K12 (Fig. 1), [22]. Another putrescine importer system, referred to as YeeF, has also been described. This system depends on proton motive forces [19].
The precise role of polyamine biosynthesis pathways and transport systems has not been studied in pathogenic E. coli. Considering that APEC strains have the ability to spread to different body sites, where intestinal E. coli do not grow, we hypothesized that polyamine biosynthesis and uptake might be essential for growth of APEC. In order to investigate whether the system described for E. coli K12 is present in APEC strains, we first performed a genomic investigation of an ST117 APEC strain. Secondly, we investigated the growth ability of polyamine biosynthesis and transport mutants of the same APEC strain (ST117- O83:H4). As a control, we compared the observed results to those obtained for a standard K-12 strain (MG1655).
Results
Characterization of the APEC strain WT-ST117 based on whole genome sequencing (WGS)
The APEC strain WT-ST117 has previously been characterized by phenotypic tests and traditional MLST typing [5] and its virulence potential has been determined [23, 24]. Before being used to analyse the importance of polyamines during growth, it was characterized by whole genome sequencing. This confirmed that the strain belongs to ST-type ST117 and serotype O83:H4 as previously described [5]. It was found to contain several virulence factors, many of which are associated with pathogenicity islands (PAI), such as papA, fimH, papEF, and ireA, as well as genes harbored by a large avian E. coli plasmid termed pAPEC-O2-ColV (iroN, sitA, iss) [24]. The gene feoB, which has been described in almost all APEC isolates [23], was also found. Other virulence genes identified are also commonly seen among APEC isolates, such as iucD and tsh (related to pAPEC-O2-ColV), as well as papC and ast, both associated to adhesin formation [24]. Furthermore, the strain was found to carry multiple iron acquisition systems, which may reflect the ability of the bacteria to grow in environments with low concentrations of iron during the infection, as described in Janßen [25]. The isolate was shown to harbour genes encoding resistance to aminoglycosides (aadA1) and sulphonamide (sul1). This genotype was in accordance with phenotypic resistance results (Additional file 1:Table S2). Besides, the WT-ST117 strain was found to contain pAPEC-O2-ColV (NC_007675) plasmid, and IncFIB/IncFII plasmid replicons.
The polyamine biosynthesis and transport genes described in MG1655 (https://biocyc.org) were conserved and all of them were present in WT-ST117. A summary of the genomic features is given in Table 1. Interestingly, WGS revealed that in the APEC strain, a small sequence encoding 51-amino acid residue of a hypothetical protein (HP), which is not present in the genome of MG1655 in the same position (NC_000913.3), was located immediately upstream of potE, as shown in (Additional file 1: Figure S1 ). This observation suggests that residues of a particular hypothetical protein may be inserted into different regions [26]. NCBI analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi#alnHdr_1379240753) and randomly selected isolates on EnteroBase (http://enterobase.warwick.ac.uk/species/ecoli/search_strains?query=st_search) indicate that this genetic configuration is observed in ExPEC isolates (NC_017659.1/NC_011751.1), but it is not commonly found among APEC isolates.
Analysis of growth in vitro
In order to investigate whether the genes involved in putrescine and spermidine biosynthesis and in transport of polyamines are required for growth in minimal medium, we analysed the lag-phase and the growth rates of biosynthesis and transport mutant strains compared to the corresponding wild type strains. Results showed that none of the MG1655 biosynthesis mutants deviated significantly from the WT strain with regards to length of lag-phase or growth rate. This is confirmation of previously published results in E. coli K12 [26, 27] (Additional file 1: Figure S2A and S2B).
A slight increase of the lag-phase was observed for ΔspeB-ST117 and its growth rate, although not significant, was below the growth rate of the WT-ST117 in minimal medium, while the mutant in speC grew exactly as the wild type strain (Fig. 2a). A significant increase of the lag-phase and a significantly reduced growth rate (μ = 0.27 compared to μ = 0.52 for WT, p = 0.009) were observed in the double biosynthesis mutant (ΔspeB/C-ST117) when grown in minimal medium (Fig. 2a) (Table 2). WGS confirmed that the ΔspeB/C mutant lacked the expected specific genes, which were replaced with selectable markers (chloramphenicol/trimethoprim), and that no additional mutations were present in the strain. The results were confirmed by basic local alignment search tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and alignment software MAUVE (http://darlinglab.org/mauve/mauve.html). Thus, every phenotype observed for the mutant could unambiguously be related to the site-specific mutations.
When the media was supplemented with putrescine or spermidine, the growth performance of the mutant was similar to that of the WT-ST117 (Fig. 2b and c), showing that extracellular polyamines could compensate for lack of biosynthesis. Construction of a triple mutant (ΔspeB/C/F-ST117) was not possible, with any of the techniques used. The remaining biosynthesis mutants grew similarly to WT-ST117 in all the media tested (Fig. 2a, b and c); thus lack of production of spermidine due to knock-out of speD together with speE was not crucial for the growth of the bacterium, even when growing in M9, which does not contain this polyamine.
Mutants of MG1655 in polyamine transport-systems grew similar to wild type MG1655 in minimal media whether supplemented with polyamines or not (Additional file 1: Figure S2B). Mutants in polyamine transport systems of ST117 also grew similar to the wild type strain, with the exception of ΔpotE-ST117, (Fig. 3a, b and c).
The ΔpotE-ST117 mutant showed an increased lag-phase and a significantly reduced growth rate in M9 medium (μ = 0.26 compared to μ = 0.52 for WT, p = 0.005) (Table 2), although it could reach the same OD as WT-ST117 after 16 h (Fig. 3a). Addition of exogenous putrescine to the medium shortened the lag-phase of the ΔpotE-ST117 mutant; however, the growth rate was still significantly different from the WT-ST117 (μ = 0.37 compared to μ = 0.46 for WT, p = 0.005) (Fig. 3b). Furthermore, it was significantly higher than in medium not supplemented with putrescine (Table 2). Complementation with the potE gene in trans (ΔpotE/pACYC184) restored the wild type phenotypes (Fig. 3a and b). Interestingly, when adding spermidine to the minimal medium, ΔpotE-ST117 grew similarly to WT-ST117 (Fig. 3c).
We also investigated the growth performance of the WT-ST117, the ΔspeB/C, ΔpotE mutants as well as the ΔpotE/pACYC184 complemented mutant in minimal medium supplemented with ornithine or arginine, since these two amino acids are putrescine precursors, and ornithine can be exchanged by the PotE antiport mechanism (Fig. 1). The addition of exogenous ornithine and arginine to the media resulted in an increase of the lag-phase of ΔspeB/C-ST117, (over 10 h delay), and still, the strain showed a significantly reduced growth rate (μ = 0.30 compared to μ = 0.41 for WT; p = 0.002) (Fig. 4a) and (μ = 0.40 compared to μ = 0.47 for WT; p = 0.03) (Fig. 4b) (Table 2), respectively. In contrast, the addition of ornithine restored ΔpotE-ST117 growth rate to the levels of WT-ST117 (Fig. 4a). The ΔpotE-ST117 mutant showed a significantly increased lag-phase and significantly reduced growth rate compared with the WT (μ = 0.28 compared to μ = 0.47 p = 0.002), when grown in M9 supplemented with arginine (Fig. 4b) (Table 2). To ensure that none of the growth results were artefacts of the small volume of media used in the Bioscreen format, the growth experiments with ornithine and arginine were repeated in 100 ml media in flasks. This showed that growth curves determined by Bioscreen automated system were in agreement with growth curves determined by the traditional cultivation method (Additional file 1: Figure S3 and Additional file 1: Table S1).
Analysis of the intracellular amount of putrescine during growth
Since the growth experiments showed that two mutants reported to be involved in intracellular putrescine homeostasis affected the growth rate of the APEC strain under study, we analysed the intracellular putrescine levels in the WT strains and in ΔspeB/C-ST117 and ΔpotE-ST117 by UHPLC during growth in minimal medium with or without supplement of putrescine. The analyses revealed that the intracellular amount of putrescine in the ΔpotE-ST117 mutant was not significantly different from the WT-ST117 strain at any of the time points tested, irrespective of the media used to analyse the growth (Fig. 5). The general trend was the detection of lower values of intracellular putrescine when the bacteria were grown in minimal medium compared with the growth in medium supplemented with putrescine. In M9, until late stationary phase, the ΔpotE-ST117 mutant showed slightly lower values compared to the WT-STT17 strain, while at late stationary phase, there was a tendency that the mutant accumulated putrescine and reached higher intracellular levels (not significant) than those found in the WT-ST117 strain (Fig. 5a). When exogenous putrescine was added to the medium, the trend was the same, but in this case, the ΔpotE-ST117 mutant showed lower intracellular levels of putrescine at late stationary phase compared to WT-ST117 (not significant). Besides, in the supplemented medium, both strains showed higher intracellular levels of putrescine at each time point tested than at the equivalent time points analysed in M9, but also these differences were not significant (Fig. 5b).
The double biosynthesis mutant, ΔspeB/C-ST117 had a remarkable lower putrescine intracellular concentration than WT-ST117 in minimal medium. The difference was significant at all-time points tested (Fig. 6a). When exogenous putrescine was added to the medium, the amount of intracellular putrescine was similar and not significantly different to the amount observed inside the WT-ST117 at all-time points (Fig. 6b).
Analysis of membrane stress caused by SDS
In order to investigate whether PotE might have a role in protection against membrane stress in APEC, as it has been described for a range of eukaryotic and prokaryotic organisms [28], we analysed if the mutant lacking potE showed increased sensitivity to SDS, which is the prototype stress factor for shock proteins [29]. The assays were performed with WT-ST117 as well as the mutant ΔpotE-ST117 and ΔpotE/pACYC184 (complemented) strains. A significantly lower CFU counts was observed for the ΔpotE-ST117 and ΔpotE-MG1655 mutants compared with their respective wild type strains when isolates were exposed to a concentration of 0.1% (w/v) SDS (Fig. 7). The CFU counts of ΔpotE-ST117 mutant were significantly lower than those observed in ΔpotE-MG1655 (Fig. 7), suggesting that this mutant was more severely affected by SDS induced stress. The ΔpotE/pACYC184 strain showed similar CFU counts to those detected for the WT strain as expected (Fig. 7). All the strains grew similarly to the WT when the medium was not supplemented with SDS or supplemented with a concentration of 0.01% (w/v) SDS (Additional file 1: Figure S4).
Gene expression of polyamine biosynthesis genes
According to the UHPLC results, the double biosynthesis mutant (ΔspeB/C-ST117) was putrescine-deficient in minimal medium, despite the presence of the speF gene. We therefore analysed whether the mutant expressed the speF gene, since this gene encodes a redundant enzyme for the same reaction where speC is involved in (Fig. 1). In addition, the expression of speE, which encodes the enzyme responsible for conversion of putrescine to spermidine (Fig. 1) was analysed. Results showed that both genes were expressed during growth in minimal medium, at all-time points tested (Additional file 1: Figure S5).
Discussion
In this study, we investigated whether genes involved in biosynthesis or transport of polyamines were required for the in vitro growth of an APEC E. coli isolate, WT-ST117, despite the fact that they are dispensable for growth in E. coli K12 [26, 27]. For this purpose, the growth performance of several mutants was compared with that of WT-ST117. Our hypothesis was that the concomitant blocking of the agmatine and the ornithine pathways would inhibit the synthesis of putrescine and would lead to a significant fitness cost in APEC E. coli when the bacteria grow under minimal conditions.
The results showed that the phenotype of a single gene knock-out was stronger (slightly attenuation of growth rate observed) for ΔspeB-ST117 than for ΔspeC-ST117, suggesting that speB is more important than speC for putrescine biosynthesis in APEC WT-ST117 and that arginine might be preferred over ornithine as the source for the biosynthesis. Furthermore, we observed that the double mutant, ΔspeB/C-ST117 showed an increased lag-phase and a significant delay of growth under minimal conditions. Addition of putrescine or spermidine to the medium allowed the mutant to grow similarly to the wild type strain, showing that uptake of putrescine from the environment could complement fully for the lack of biosynthesis. Spermidine was apparently not essential for growth since the double mutant without speD and SpeE grew similarly to the wild type strain. Curiously, the addition of spermidine to the medium compensated for the lack of putrescine biosynthesis. This suggests that spemidine is taken up and then converted by either speE or other enzymes to putrescine, in the lack of putrescine biosynthesis. This has not previously been described in bacteria, however, it is well known from eukaryotic polyamine biosynthesis systems that spermidine can be converted to putrescine [30]. On the other hand, conversion of spermidine to putrescine in the absence of an external source of spermidine was not sufficient to compensate the lack of putrescine production, and further studies are warranted to understand the regulation of this system.
Similar study in Streptococcus pneumoniae polyamine-deficient mutant pointed that putrescine biosynthesis is crucial for fitness [16]. However, previous studies with E. coli-HT306 [26] and similar work carried out on the close relative Salmonella [11], demonstrated that polyamines are not required for growth in vitro, although polyamines were shown to be essential for systemic infection of mice with Salmonella [31]. Thus, despite the close genetic relationship to other strains of E. coli and to Salmonella, it seems that deletions in the polyamines biosynthesis pathways in the pathogenic APEC E. coli WT-ST117 strain is crucial to growth, and it may indicate that polyamines play a different role during APEC (ST117- O83:H4) growth than the described in these other bacteria.
We measured the amount of intracellular putrescine inside WT-ST117 and the isogenic strain ΔspeB/C-ST117 at different phases of growth by UHPLC and found that the amount of putrescine within WT-ST117 changed over time, increasing with growth, probably due to the bacterial multiplication. The double biosynthesis mutant was clearly putrescine-deficient and only very low levels of putrescine were detectable at just one-time point. This finding shows that the double biosynthesis mutant does not generate putrescine during growth in minimal medium, or the amounts of synthesized putrescine are not detectable by UHPLC.
Theoretically, the ΔspeB/C-ST117 mutant should still have been able to synthesize putrescine from ornithine because the speF gene remains present. This was not the case, and we observed that addition of ornithine to the medium, which is the substrate for speF, did not restore the growth defect. We cannot fully rule out that the lack of putrescine in this mutant, despite the presence of SpeF, might be due to continuous conversion of putrescine into spermidine catalyzed by SpeE. However, we consider this an unlikely explanation since spermidine appeared to be less important to the cell than putrescine. Nevertheless, under the growth conditions tested, both speF and speE were expressed in ΔspeB/C-ST117.
We also observed that the mutant lacking the gene ΔpotE, involved in putrescine homeostasis through transport of putrescine and ornithine, showed an increased lag-phase and significantly reduced the growth rate in minimal medium. It was also growth attenuated in minimal medium supplemented with arginine. The amino acid L-arginine has been described as an inhibitor of bacterial proliferation at certain concentrations due to the effects on bacteria cell membrane leading to disruption and depolarization [32]. This may partly explain the growth arrest in the medium where this amino acid was added. In contrast, the addition of spermidine or ornithine to the media, allowed the mutant to grow similarly to the WT-ST117 strain.
Our first hypothesis to explain the growth attenuation of this mutant was that the lack of the transport system, PotE, might lead to intracellular accumulation of putrescine. Such an explanation would be consistent with the fact that the PotE protein has been described as the main protein associated with the excretion of putrescine [21]. However, our results demonstrated that such intracellular accumulation of putrescine did not take place in any of the media tested; actually, putrescine was found inside of the mutant at lower levels than those observed inside of WT-ST117, even when grown in M9 medium supplemented with putrescine. Therefore, putrescine might either be produced in balanced levels eliminating the need for excretions, or it may be excreted through another transport system or metabolized with other purposes in the mutant. Our findings suggest that the function of PotE may be wider than previously described [22, 34], and since ornithine and spermidine added to the medium eliminated the growth attenuation, it may be an uptake system for these substances; however, it was considered beyond the scope of the current work to investigate the uptake and export repertoire of this molecule.
Reactive oxygen species (ROS) are formed as part of normal cell metabolism. In mammals, oxidation of polyamines can generate highly toxic products such as acrolein, which causes oxidative damage to the cells [30]. In yeast cells, it has been described that central components of the stress response are regulated by polyamine exporter systems [33]. The lack or the excess of polyamines can induce oxidative stress in E. coli; the increase of oxidant levels are associated with changes in the metabolism and cell cycle arrests [34, 35]. In light of this, our second hypothesis was that the absence of the transport system (PotE) might lead to a decreased ability to cope with oxidative stress and that the bacterial cells react to the increased oxidative level by arresting the growth. Notably, our findings suggest that PotE has a protective role against oxidative stress in the E. coli APEC strain since the mutant ΔpotE-STT17 was vulnerable to SDS induced membrane stress. Interestedly, the stress oxidase induction seems to be critical in the pathogenic strain. This would also explain with addition of arginine increased the growth attenuation of the ΔpotE mutants, since this molecule may add to membrane stress [33]. Therefore, the absence of the PotE membrane protein may compromise the pathogenic bacteria survival under stressful conditions.
Conclusions
Putrescine biosynthesis was found to be important for APEC (ST117- O83:H4) when growing in minimal medium, a phenotype which differs from the phenotype previously described for other types of E. coli [26]. Also the PotE transport system was found to be important for growth. One function of polyamines is to maintain the charge balance of the cell and act as a shield stabilizing the cell membrane [36, 37]. Considering that PotE is a putrescine membrane exporter [22], our results indicate that the growth arrest observed for this mutant is a response to oxidative stress. Thus, we suggest that the PotE membrane protein plays a different role during growth of the pathogenic APEC E. coli (ST117- O83:H4) strain than during growth of standard laboratory strains, or that these strains have additional systems to cope with membrane stress caused by polyamine imbalance. Despite numerous attempts, we were not able to obtain mutants lacking both potE (involved in transport of putrescine) and speB/C (required for biosynthesis of putrescine) in our APEC strain (ST117- O83:H4). This suggests that at least one of the systems needs to be intact for any growth to occur.
Methods
Bacterial strains and growth conditions
Experiments were performed with an APEC E. coli wild type strain O83:H4 (WT-ST117) [5], and E. coli MG1655 (K-12) (MG1655) from the strain collection at Department of Veterinary and Animal Sciences, University of Copenhagen. Mutants of these two strains were created as part of the current study (see below). The WT-ST117 was originally isolated from chronic lesions of salpingo-peritonitis in a broiler breeder in Denmark, and belongs to serotype O83:H4, sequence type ST117 and phylogroup D [5]. Prior to growth experiments, bacterial strains were propagated for 16 to 24 h at 30–37 °C in Luria-Bertani (LB) broth (Oxoid) with continuous shaking (200 rpm), or they were cultured overnight on LB agar plates, supplemented with gentamicin 20 μg/ml (Sigma-Aldrich), trimethoprim 10 μg/ml (Sigma-Aldrich) and/or chloramphenicol 5 μg/ml (Sigma-Aldrich) when relevant.
Mutagenesis
Single gene deletions and concomitant insertion of antibiotic resistance cassettes in WT-ST117 and MG1655 were performed using Lambda Red recombination [38], with the exception of the double mutant ∆speB/C, which was obtained by using the In-fusion HD method (Clontech). Genetic complementation of potE in WT-ST117 was achieved by cloning the potE gene into the plasmid pACYC184 using standard techniques. The resulting plasmid was electroporated into E. coli K-12 (ER2420), from which it was recovered and eventually electroporated into the ΔpotE-ST117 mutant as described [39]. Mutated strains and plasmids used for mutagenesis are listed in Table 3, while primers used for amplification of the genes are listed in (Additional file 1: Table S3). All constructs were verified by PCR amplification, restriction analysis and sequencing.
Whole genome sequencing
Genomic DNA was extracted from WT-ST117 and its isogenic ∆potE-ST117, ΔspeD/E-ST117, and ∆speB/C-ST117 strains using the Invitrogen Easy-DNATM Kit (Invitrogen) as recommended by the supplier. DNA concentrations were determined by the Qubits DNA BR assay Kit (Invitrogen). The genomic DNA was prepared for Illumina pair-end sequencing using the Illumina (CD genomics) NexteraXT Guide 150,319,425,031,942 following the protocol revision C (https://support.illumina.com/downloads/nextera_xt_sample_preparation_guide_15031942.html). A sample of the pooled NexteraXT Libraries was loaded onto an Illumina MiSeq reagent cartridge by MiSeq Reagent Kit v2 and 500 cycles with a Standard Flow Cell. The libraries were sequenced using an Illumina platform MiSeq v2.6. The isolates were pair-end sequenced. The raw reads were assembled and contigs aligned using CLC Workbench software (CLC Bio-Qiagen). The assembled genome of WT-ST117 and polyamine mutants was submitted to European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) under study accession number ‘PRJEB18262’ and the samples accession numbers are ‘ERS1461989’ (WT-ST117), ‘ERS2614665’ (ΔspeD/E-ST117), ‘ERS2614666’ (∆speB/C-ST117), ‘ERS2614667’ (∆potE-ST117).
The assembled sequences were analyzed using the pipelines available at the Centre for Genomic Epidemiology (CGE; http://www.genomicepidemiology.org); KmerFinder (version 2; https://cge.cbs.dtu.dk/services/KmerFinder/), and SeroTypeFinder (version 1.1; https://cge.cbs.dtu.dk/services/SerotypeFinder). Other genotypic features such as plasmid replicons, antimicrobial resistance genes, and virulence genes were also identified with a selected threshold equal to 98% identity using the pipelines; PlasmidFinder (version 1.3; https://cge.cbs.dtu.dk/services/PlasmidFinder) ResFinder (version 2.1; https://cge.cbs.dtu.dk/services/ResFinder) and VirulenceFinder (version 1.4; https://cge.cbs.dtu.dk/services/VirulenceFinder) also available at CGE. Specific genes, such as those of the polyamine biosynthesis and transport systems were identified by Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) using the sequence of E.coli MG1655 (https://www.ncbi.nlm.nih.gov/nuccore/NC_000913.3) as the reference. The sequences of ∆potE-ST117 and ∆speB/C-STS117 were compared to that of WT-ST117 to confirm that gene knock-outs were site-specific.
In vitro growth assays
Growth experiments were performed in biological triplicate on BioScreenC (Labsystems) for 24 h at 37 °C. Bacteria were grown overnight in M9-minimal medium (2 mM MgSO4, 0.1 mM CaCl2, 0.4% glucose, 8.5 mM NaCl, 42 mM Na2HPO4, 22 mM KH2PO4, and 18.6 mM NH4Cl) and adjusted to the same OD (OD600 = 0.05). A volume of 190 μl from each medium tested; M9 and M9 supplemented with putrescine 1.13 mM or spermidine 0.07 mM was added in the honeycomb Bioscreen wells and 10 μl of the bacterial suspension was inoculated. The OD values (recorded with a 600 nm filter) were measured every 15 min with continuous shaking at 37 °C. In addition, the medium was added to several wells and used as blank control. Growth curves were created on the basis of a non-linear model of the log-transformed OD600 values with the definition of lag-phase as the time necessary to reach an OD600 of 0.1. The maximum specific growth rate (μ) was calculated according to the logistic equation model using the biological triplicate values [40].
In addition, biological triplicate growth assays of WT-ST117, ΔpotE-T117, ΔspeB/C-ST117 and ΔpotE/pACYC184 (complemented) strains were performed in the M9 supplemented with ornithine 0.7 mM or arginine 0.7 mM, under the same conditions. For the standard cultivation method, bacteria were inoculated in 100 ml media in flasks with M9-minimal medium, M9 supplemented with putrescine, spermidine, ornithine, or arginine with continuous shaking at 37 °C. At each time point (30 min intervals), OD600 values were checked out.
Ultra-high performance liquid chromatography (UHPLC)
Intracellular concentrations of putrescine in the strains; WT-ST117, ΔpotE-ST117 and ΔspeB/C-ST117 were estimated by UHPLC. Samples were harvested at five different time points during growth in M9 and M9 supplemented with putrescine; T0- initial lag-phase (OD600 = 0.05), T1- start of the logarithmic phase (OD600 = 0.5), T2- mid-logarithmic phase (OD600 = 1), T3- start of the stationary phase (OD600 = 1.8–2), and T4- late stationary phase (OD600 = 3–4). The samples were centrifuged and washed with sterile PBS and immediately frozen and stored at − 20 °C.
For putrescine determination, the frozen cellular pellets were resuspended in 500 μL of 0.1 N HCl and boiled for 15 min. Then, a volume of 100 μL of the mixture was derivatized with diethyl ethoxymethylenemalonate (DEEMM) and separated by UHPLC following the protocol described in Redruello et al. [41]. Data were acquired and analyzed using Empower 2 software (Waters). All the chemicals used were of the highest available purity. The water of Milli-Q quality (Millipore) was used in all solutions. HPLC-grade acetonitrile (VWR) and methanol (Merck) were used as pure solvents. The putrescine concentration in total cellular homogenates is expressed as mg/L.
Induction of membrane stress caused by SDS
The WT-ST117, the mutant ΔpotE-ST117, and the complemented mutant ΔpotE/pACYC184-ST117 strains were grown overnight in LB at 37 °C, 200 rpm, adjusted to an OD600 = 1, and 10-fold dilutions were prepared. 100 μl of each dilution were cultured on LB agar plates and LB agar plates supplemented with 0.01% (w/v) and 0.1% (w/v) SDS (Sodium dodecyl sulfate) (Thermo Scientific). CFU counts were assessed after 16–18 h of incubation at 37 °C. The experiment was performed in triplicate.
RNA extraction and qPCR
To analyze the gene expression level of the genes speF and speE in the mutant ∆speB/C-STS117, bacteria were harvested at three different time points during growth in M9; start of the logarithmic phase, mid-logarithmic phase and stationary phase (OD600 = 0.5, OD600 = 1 and OD600 = 2, respectively). RNA was isolated from the cultures (1.5 ml) by mechanical disruption with the FastPrep (Bio101; Qbiogene) and RNeasy mini kit (Quiagen). Quantity and quality of total RNA were determined with the NanoDrop 1000 spectrophotometer (Thermo Scientific) and on a 1.5% (w/v) agarose gel. To remove chromosomal DNA, RNA samples were treated with RNase free DNase I kit (Thermo Scientific). The cDNAs were synthesized using the GoScript reverse transcriptase kit (Promega). The qPCR was done using the Maxima SYBR Green/Rox qPCR Master Mix and gene-specific oligonucleotides, which are described in the (Additional file 1: Table S3), in a LightCycler 96 (Roche). Data were normalized against the reference gene 16S rRNA [42] and normalization was determined based on the established mathematical model [43]. The experiment was performed in triplicate.
Statistical analysis
Statistical analysis was performed using the GraphPad Prism (GraphPad Software) version 5.0. Differences between the mean growth rate of strains and between the amounts (mg/L) of putrescine in different strains were evaluated by students t-test. A P-value of < 0.05 was considered significant.
Abbreviations
- Amp:
-
Ampicillin
- APEC:
-
Avian pathogenic Escherichia coli
- ATCC:
-
American type culture collection
- BLAST:
-
Basic local alignment search tool
- CFU:
-
Colony formation unit
- CGE:
-
Center for Genomic Epidemiology
- Chl:
-
Chloramphenicol
- ENA:
-
European nucleotide archive
- Gen:
-
Gentamicin resistant
- HPLC:
-
High performance liquid chromatography
- LB:
-
Luria Bertani
- M9:
-
Minimal medium
- MAUVE:
-
Multiple genome alignment
- OD:
-
Optical density
- PAI:
-
Pathogenicity islands
- PBS:
-
Phosphate-buffered saline
- PEG:
-
Promoter extraction
- qPCR:
-
quantitative PCR
- ROS:
-
Reactive oxygen species
- rpm:
-
revolution per minute
- SDS:
-
Sodium dodecyl sulphate
- Tet:
-
Tetracycline
- Tmp:
-
Trimethoprim
- UHPLC:
-
Ultra-high performance liquid chromatography
- VWR:
-
Van waters and Rogers
- w/v :
-
weight per volume
- WGS:
-
Whole genome sequencing
- WT:
-
Wild type
References
Johnson JR, Russo TA. Molecular epidemiology of extraintestinal pathogenic (uropathogenic) Escherichia coli. Int J Med Microbiol. 2005;295:383–404.
Mora A, López C, Herrera A, Viso S, Mamani R, Dhabi G, et al. Emerging avian pathogenic Escherichia coli strains belonging to clonal groups O111: H4-D-ST2085 and O111: H4-D-ST117 with high virulence-gene content and zoonotic potential. Vet Microbiol. 2012;156:347–52.
Van Der Westhuizen WA, Bragg RR. Multiplex polymerase chain reaction for screening avian pathogenic Escherichia coli for virulence genes. Avian Pathol. 2012;41:33–40.
Ronco T, Stegger M, Olsen RH, Sekse C, Nordstoga AB, Pohjanvirta T, et al. Spread of avian pathogenic Escherichia coli ST117 O78:H4 in Nordic broiler production. BMC Genomics [Internet]. BMC Genomics; 2017;18:13.
Pires-dos-Santos T, Bisgaard M, Christensen H. Genetic diversity and virulence profiles of Escherichia coli isolates causing salpingits and peritonitis in broiler breeders. Submitted for publication. 2013;162:873–80.
Lee J, Sperandio V, Frantz DE, Longgood J, Camilli A, Phillips MA, et al. An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in Vibrio cholerae. J Biol Chem. 2009;284:9899–907.
Miller-Fleming L, Olin-Sandoval V, Campbell K, Ralser M. Remaining Mysteries of Molecular Biology: The Role of Polyamines in the Cell. J Mol Biol. 2015;427:3389–406.
Kurihara S, Oda S, Kato K, Kim HG, Koyanagi T, Kumagai H, et al. A novel putrescine utilization pathway involves γ-glutamylated intermediates of Escherichia coli K-12. J Biol Chem. 2005;280:4602–8.
Igarashi K, Kashiwagi K. Polyamines: mysterious modulators of cellular functions. Biochem Biophys Res Commun. 2000;271:559–64.
Joshi GS, Spontak JS, Klapper DG, Richardson AR. Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol Microbiol. 2011;82:9–20.
Jelsbak L, Thomsen LE, Wallrodt I, Jensen PR, Olsen JE. Polyamines are required for virulence in Salmonella enterica serovar typhimurium. PLoS One. 2012;7:1–10.
Schroll C, Christensen JP, Christensen H, Pors SE, Thorndahl L, Jensen PR, et al. Polyamines are essential for virulence in Salmonella enterica serovar Gallinarum despite evolutionary decay of polyamine biosynthesis genes. Vet Microbiol. 2014;170:144–50.
Nasrallah GK, Riveroll AL, Chong A, Murray LE, Lewis PJ, Garduño RA. Legionella pneumophila requires polyamines for optimal intracellular growth. J Bacteriol. 2011;193:4346–60.
Russo BC, Horzempa J, O’Dee DM, Schmitt DM, Brown MJ, Carlson PE, et al. A Francisella tularensis locus required for Spermine responsiveness is necessary for virulence. Infect Immun. 2011;79:3665–76.
Barbagallo M, Di Martino ML, Marcocci L, Pietrangeli P, de Carolis E, Casalino M, et al. A new piece of the Shigella pathogenicity puzzle: spermidine accumulationby silencing of the speG gene. PLoS One. 2011;6:e27226.
Shah P, Nanduri B, Swiatlo E, Ma Y, Pendarvis K. Polyamine biosynthesis and transport mechanisms are crucial for fitness and pathogenesis of Streptococcus pneumoniae. Microbiology. 2011;157:504–15.
Jelsbak L, Hartman H, Schroll C, Rosenkrantz JT, Lemire S, Wallrodt I, et al. Identification of metabolic pathways essential for fitness of Salmonella typhimurium in vivo. PLoS One. 2014;9:e101869.
Potter AJ, Paton JC. Spermidine biosynthesis and transport modulate pneumococcal autolysis. J Bacteriol. 2014;196:3556–61.
Kurihara S, Suzuki H, Oshida M, Benno Y. A novel putrescine importer required for type 1 pili-driven surface motility induced by extracellular putrescine in Escherichia coli K-12. J Biol Chem. 2011;286:10185–92.
Schneider J, Wendisch VF. Biotechnological production of polyamines by Bacteria: recent achievements and future perspectives. Appl Microbiol Biotechnol. 2011;91:17–30.
Shah P, Swiatlo E. A multifaceted role for polyamines in bacterial pathogens. Mol Microbiol. 2008;68:4–16.
Kashiwagi K, Kuraishi A, Tomitori H, Igarashi A, Nishimura K, Shirahata A, et al. Identification of the putrescine recognition site on polyamine transport protein PotE. J Biol Chem. 2000;275:36007–12.
Zhao L, Gao S, Huan H, Xu X, Zhu X, Yang W, et al. Comparison of virulence factors and expression of specific genes between uropathogenic Escherichia coli and avian pathogenic E. coli in a murine urinary tract infection model and a chicken challenge model. Microbiology. 2009;155:1634–44.
Rodriguez-Siek KE, Giddings CW, Doetkott C, Johnson TJ, Nolan LK. Characterizing the APEC pathotype. Vet Res. 2005;36:241–56.
Jansen T. Virulence-associated genes in avian pathogenic (APEC) isolated from internal organs of poultry having died from colibacillosis. Int J Med Microbiol. 2001;291:371–8.
De Las RB, Marcobal A, Muñoz R. Gene organization of the ornithine decarboxylase-encoding region in Morganella morganii. J Appl Microbiol. 2007;102:1551–60.
Chattopadhyay MK, Tabor CW, Tabor H. Polyamines are not required for aerobic growth of Escherichia coli: Preparation of a strain with deletions in all of the genes for polyamine biosynthesis. J Bacteriol. 2009;191:5549–52.
Igarashi K, Kashiwagi K, Hamasaki H, Miura A, Kakegawa T, Hirose S, et al. Formation of a compensatory polyamine by Escherichia coli polyamine-requiring mutants during growth in the absence of polyamines. J Bacteriol. 1986;166:128–34.
Rhee HJ, Kim EJ, Lee JK. Physiological polyamines: Simple primordial stress molecules. J Cell Mol Med. 2007;11:685–703.
Pegg AE. Mammalian polyamine metabolism and function. IUBMB Life. 2009;61:880–94.
Cartas I, Regina P, Jelsbak L. Microbial Pathogenesis Multiple roles of putrescine and spermidine in stress resistance and virulence of Salmonella enterica serovar Typhimurium. Microb Pathog. 2016;95:117–23.
Rodríguez E, Seguer J, Rocabayera X, Manresa A. Cellular effects of monohydrochloride of L-arginine, Nα- lauroyl ethylester (LAE) on exposure to Salmonella Typhimurium and Staphylococcus aureus. J Appl Microbiol. 2004;96:903–12.
Krüger A, Vowinckel J, Mülleder M, Grote P, Capuano F, Bluemlein K, et al. Tpo1-mediated spermine and spermidine export controls cell cycle delay and times antioxidant protein expression during the oxidative stress response. EMBO Rep. 2013;14:1113–9.
Chattopadhyay MK, Tabor CW, Tabor H. Polyamines protect Escherichia coli cells from the toxic effect of oxygen. Proc Natl Acad Sci. 2003;100:2261–5.
Tkachenko A, Nesterova L. The role of the natural polyamine putrescine in defense against oxidative stress in Escherichia coli. 2001;:155–7.
Gupta K, Sengupta A, Chakraborty M, Gupta B. Hydrogen Peroxide and Polyamines Act as Double Edged Swords in Plant Abiotic Stress Responses. Front Plant Sci. 2016;7:1343.
Schuber F. Influence of polyamines on membrane functions. Biochem J. 1989;260:1–10.
Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. PNAS. 2000;97:6640–5.
Terui Y, Saroj SD, Sakamoto A, Yoshida T, Higashi K, Kurihara S, et al. Properties of putrescine uptake by PotFGHI and PuuP and their physiological significance in Escherichia coli. Amino Acids. 2014;46:661–70.
Mytilinaios I, Salih M, Schofield HK, Lambert RJW. Growth curve prediction from optical density data. Int J Food Microbiol. 2012;154:169–76.
Redruello B, Ladero V, Cuesta I, Álvarez-Buylla JR, Martín MC, Fernández M, et al. A fast, reliable, ultra high performance liquid chromatography method for the simultaneous determination of amino acids, biogenic amines and ammonium ions in cheese, using diethyl ethoxymethylenemalonate as a derivatising agent. Food Chem. 2013;139:1029–35.
Gao W, Zhang W, Meldrum DR. RT-qPCR based quantitative analysis of gene expression in single bacterial cells. J Microbiol Methods. 2011;85:221–7.
Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25.
Bossé JT, Li Y, Walker S, Atherton T, Crespo RF, Williamson SM, et al. Identification of dfrA14 in two distinct plasmids conferring trimethoprim resistance in Actinobacillus pleuropneumoniae. J Antimicrob Chemother. 2015;70:2217–22.
Acknowledgments
The authors would like to thank Fabio Antenucci for his help to construct the double ΔspeB/C-ST117 mutant; Egle Kudirkiene and Steffen L. Jørgensen are thanked for bioinformatics advice and Kaisong Huang for qPCR assistance.
Funding
Priscila R. Guerra was supported by a scholarship from CAPES – Brazilian Federal Agency for Support and Evaluation of Graduate Education within the Ministry of Education of Brazil. This study received financial support from Spanish Ministry of Economy, Industry and Competitiveness (AGL2013–45431-R and AGL2016–78708-R), and from the Danish Council for Independent Research (Technology and Production), grant number DFF – 4184-00050. The sponsors had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in ENA repository, https://www.ebi.ac.uk/ena/data/view/PRJEB18262.
Author information
Authors and Affiliations
Contributions
PRG designed the study, performed all experiments and analyzed results; MRS performed the growth experiment; VL and BR performed the UHPLC analysis; TPS contributed to the design of the study; AHF, LJ, and JEO designed the study, participated in analysis of results, and supervised the study. PRG wrote the first draft of the manuscript and all authors read and approve the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S1-S5. and Tables S1-S3. Description of Data: The additional file contains information on the genomic organization of potE in E. coli WT-ST117 and E. coli MG1655, growth phenotypes of polyamine biosynthesis mutants, growth performance of the mutants under stress conditions, expression levels of polyamine biosynthesis genes, correlation between CFU counting and OD values, antimicrobial phenotype results, and oligonucleotide sequences for PCR-based amplification. (PDF 614 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Guerra, P.R., Herrero-Fresno, A., Ladero, V. et al. Putrescine biosynthesis and export genes are essential for normal growth of avian pathogenic Escherichia coli. BMC Microbiol 18, 226 (2018). https://doi.org/10.1186/s12866-018-1355-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-018-1355-9