Abstract
Background
DNA methylation is a crucial epigenomic mechanism in various biological processes. Using whole-genome bisulfite sequencing (WGBS) technology, methylated cytosine sites can be revealed at the single nucleotide level. However, the WGBS data analysis process is usually complicated and challenging.
Results
To alleviate the associated difficulties, we integrated the WGBS data processing steps and downstream analysis into a two-phase approach. First, we set up the required tools in Galaxy and developed workflows to calculate the methylation level from raw WGBS data and generate a methylation status summary, the mtable. This computation environment is wrapped into the Docker container image DocMethyl, which allows users to rapidly deploy an executable environment without tedious software installation and library dependency problems. Next, the mtable files were uploaded to the web server EpiMOLAS_web to link with the gene annotation databases that enable rapid data retrieval and analyses.
Conclusion
To our knowledge, the EpiMOLAS framework, consisting of DocMethyl and EpiMOLAS_web, is the first approach to include containerization technology and a web-based system for WGBS data analysis from raw data processing to downstream analysis. EpiMOLAS will help users cope with their WGBS data and also conduct reproducible analyses of publicly available data, thereby gaining insights into the mechanisms underlying complex biological phenomenon. The Galaxy Docker image DocMethyl is available at https://hub.docker.com/r/lsbnb/docmethyl/.
EpiMOLAS_web is publicly accessible at http://symbiosis.iis.sinica.edu.tw/epimolas/.
Similar content being viewed by others
Background
DNA methylation on cytosine is an epigenetic modification that occurs in numerous biological processes, including transposable element silencing, mammalian gene regulation, genomic imprinting, and X chromosome inactivation [1]. Compared to other epigenetic modifications, cytosine methylation is a relatively stable epigenetic mark inherited during cell divisions. In vertebrates, methylated cytosines were first identified in gene promoters as well as the transcribed regions. Besides those found in promoters, the methylation pattern in intragenic transcribed gene body regions are also evolutionarily conserved among organisms, and the control and biological mechanisms remain to be explored [2].
Over the past decades, several protocols and assays, such as MBD-seq [3], MeDIP-seq [4], reduced representation bisulfite sequencing (RRBS) [5], whole-genome bisulfite sequencing (WGBS) [6], and Infinium Methylation 450 K/EPIC array [7], have been developed to profile genome-wide DNA methylation. MBD-seq and MeDIP-seq techniques are affinity enrichment-based methods that use antibodies to extract the methylated genomic regions. They are cost-effective approaches but have a potentially confounding bias in varying CpG density. Several pipelines are available and used for these type of datasets [8,9,10,11].
Bisulfite sequencing (BS-seq) has become a popular technology to analyze DNA methylation. It is based on the differential chemical reactions to bisulfite treatment between unmethylated and methylated cytosines. The treatment of sodium bisulfite converts unmethylated cytosines (C) to uracils (U) and uracils (U) to thymines (T) after PCR amplification [12]. Coupled with next-generation sequencing technology and downstream bioinformatics strategies, the bisulfite-converted reads are mapped using a wild-card or three-letter mapping strategy [13], after which, the percentage of cells that are methylated at each genomic cytosine site can be estimated. Recent advances in sequencing technology make it possible to identify the methylation states on a genome-wide scale at single-base resolution and allow WGBS data to be more accessible. Here, we focused our analysis on WGBS data. Although many approaches and tools are available to handle WGBS data [13,14,15], there is still a lack of automated workflows and well-annotated databases with customizable downstream analyses for the users’ own datasets.
Implementation
By leveraging the Linux container virtualization technology (LXC) and the community-supported Galaxy platform [16], we developed a seamless and ready-to-use workflow, which can be rapidly deployed and executed on a single machine or a distributed cloud computing environment, avoiding tedious software installation and library dependency problems. This Galaxy Docker container, DocMethyl, includes FastQC [17], Trim Galore [18], Bismark [19], the in-house program EpiMolas.jar [20], and two built-in workflows to streamline each processing step, including (1) clean-up, to trim the adapter sequence and low-quality bases from raw reads; (2) read mapping, to align trimmed reads to the reference genome; (3) methylation calling, to extract the methylation status of each cytosine throughout the genome; and (4) methylatn scoring, to calculate the methylation level of each gene.
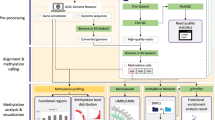
EpiMOLAS_web is an online platform that connects the DocMethyl summary file, mtable, to a rich collection of annotations of the human genome (GRCh37, GRCh38), the mouse genome (GRCm38), and the Arabidopsis genome (TAIR10) [21]. We implemented the system using the Ubuntu 14.04, Apache 2.04, PostgreSQL 9.1, PHP 5.1, and Bootstrap responsive web design (RWD) framework to create a user-friendly interface. Data integration and score calculation in the EpiMOLAS_web analysis process are implemented in Java, Python, and PHP. Moreover, to help users understand their results, several data visualization tools are integrated into the system. For example, we combined Biodalliance (version 0.13.7) [22], Clustergrammer [23] and Circos (version 0.69) [24] allowing the users to browse the coordination of genes on the chromosomes and display the gene-based DNA methylation profiles. In the enrichment analysis, the gene function annotations, including Gene Ontology (GO) terms [25] and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database [26], were collected in Feb. 2019. We also utilized the BioGRID protein interaction data (version 3.5.170) [27] and implemented a dedicated protein interaction network viewer in JavaScript, Cytoscape.js [28], HTML5, and MySQL for the topological data analyses and visualization. The integrated architecture of DocMethyl and EpiMOLAS_web is depicted in Fig. 1.
Results
To alleviate the burden of BS-seq data processing and analysis, we developed EpiMOLAS, a two-phase approach which consists of DocMethyl and EpiMOLAS_web. The Docker container, DocMethyl, completes the intensive short reads processing tasks and generates a tab-delimited methylation summary file (namely mtable) for each WGBS dataset. The online web server, EpiMOLAS_web, links the output mtable files with gene annotation databases and provides versatile downstream analyses, as shown in Fig. 2.
EpiMOLAS, including DocMethyl and EpiMOLAS_web, is a two-phase approach for WGBS data analysis. DocMethyl requires Bisulfite-Seq read data, reference genome sequences, and gene annotation files to determine the DNA methylation status. The resulting mtable files are uploaded to the EpiMOLAS_web server via the web user interfaces for users to perform various types of data analyses and visualization modules
DocMethyl
To run DocMethyl, users can directly pull down the image in a Docker runtime environment (Additional file 1 Sec. 1.2). Once DocMethyl is launched, Galaxy is automatically deployed, and the service is accessible through the browser-based interface. Users upload WGBS raw reads, the reference genome, and the gene annotation file for the corresponding inputs in the workflow DocMethyl-SE or DocMethyl-PE (Fig. 3 and Additional file 1 Sec. 1.4, 1.5). The output of the workflows is a tab-delimited text file, i.e. mtable, containing the scores for the gene-based cytosine methylation level regarding the sequence context (CG, CHG, or CHH) in the promoter and gene body regions. This seven-column data format is compatible with EpiMOLAS_web, enabling users to link the methylation level with the web server. One mtable file is generated via the workflow from one WGBS dataset. Accordingly, multiple mtable files for a multi-group experiment design with control conditions and experimental conditions can be uploaded under the guidance of the EpiMOLAS_web data submission process.
EpiMOLAS_web
EpiMOLAS_web provides an arithmetic calculation of the methylation level based on the experimental conditions to identify differentially methylated genes or promoters on a particular sequence context. Furthermore, several modules are available for retrieving the methylation measures of genes, such as a full-text keyword search on the Ensembl Gene ID, gene symbol, gene description, KEGG pathway name, or a batch query by Gene IDs and gene symbols (Fig. 4). The protein interaction network, hierarchical clustering heatmap, Venn diagram and Circos plot visualization modules allow users to investigate the selected gene lists in various respects. To identify the likely biological progression, the gene lists from these data retrieval approaches are used to perform GO term analysis or KEGG pathway enrichment analysis. Further details regarding DocMethyl and EpiMOLAS_web can be found in the Additional file 1, the DocMethyl Docker Hub repository page (https://hub.docker.com/r/lsbnb/docmethyl/), and the EpiMOLAS_web (http://symbiosis.iis.sinica.edu.tw/epimolas/).
Gene set analysis and visualization
In research, the biological functions and potential mechanisms for a particular set of genes are of particular interest. Genes that associate together, therefore, may play an essential role in specific biological processes. For this reason, a gene list can be obtained from various quantitative selecting scenarios based on methylation level or from expert manually curated databases. In EpiMOLAS_web, we developed several modules for gene set enrichment analysis and visualization, such as KEGG pathway and GO term enrichment, histograms and boxplots of methylation levels, Venn diagrams, Circos plots, hierarchical clustering heatmaps, and protein-protein interaction networks (PPIN).
GO terms and KEGG pathway enrichment analysis
A set of genes of interest is usually assumed to be involved or activated in response to perturbations in specific biological processes. Through GO terms and KEGG pathway enrichment analysis, users can determine which biological processes, molecular functions, cellular components, or KEGG pathways appear to be specifically involved and have been studied in diseases, for the gene set of interest. For the enrichment score, we calculate the p-value based on a hypergeometric test [20].
PPIN analysis
We implemented a PPIN viewer to integrate, visualize, and analyze gene list members in the protein network context in the system based on Cytoscape.js, which supports network graph drawing with a force-directed layout and perturbation resilience. There are three features regarding the depiction and analysis of the interaction network: (a) search: users can search and locate the genes on the network subgraph using the gene symbols; (b) layout: the protein network layout can be displayed in Grid, Random, CoSE, Concentric, Breadthfirst, Arbor, Cola, Dagre, and Spread and with several extra network topology measures such as degree centrality, degree centrality normalized, closeness centrality, closeness centrality normalized, and betweenness centrality for network representation; (c) export: the selected gene list or network can be exported into a Cytoscape JSON file, a text file of binary protein interactions, or an image in PNG and JPG format.
Hierarchical clustering heatmap
The hierarchical clustering heatmap is a common unsupervised approach to show differential gene expression results. It is also a widely used visualization for displaying a table of numbers representing gene expression or methylation level. We integrated an interactive clustered heatmap visualization tool, Clustergrammer, into the system to show clusters in the methylation level of genes among samples.
Venn diagram and Circos plot
A Venn diagram is a simple but effective and intuitive way to examine the overlap between lists of genes. This visualization module computes the intersection of up-to four gene sets and allows users to store the results. We also integrated the Circos plot visualization module to show the location (the chromosomal coordination) of the genes in the selected list(s). This circular genome data visualization provides a different perspective of the spatial characteristics in DNA methylation across genomic regions.
Discussion
The combination of DocMethyl and EpiMOLAS_web offers an integrated solution without tedious software installation and database management. Comparisons among several well-known platforms and tools for genome-wide DNA methylation analysis, such as BAT [29], ENCODE-WGBS [30], snakePipe [31], NGI-MethylSeq [32], Mint [33], MethylPipe [34], MethylSig [35], and Methylkit [36] with EpiMOLAS are presented in Table 1. BAT, NGI-MethylSeq and EpiMOLAS use Docker containerization technology to allow fast and simple environment deployment. Apart from EpiMOLAS and Mint, the other platforms are executed in shell scripts or workflow management systems, lacking a user-friendly web interface to satisfy the needs of laboratory researchers. Mint is accessible ins both the command line and Galaxy graphical user interface; however, it requires additional efforts to install tools and to customize the environment. Tools including ENCODE-WGBS, snakePipe, and NGI-MethylSeq have been developed to support analyses of epigenetic profiling and other -omics data (RNA-seq, ChIP-seq, Hi-C, ATAC-seq, and etc.). Nevertheless, it remains challenging to handle different -omics profiles owing to the lack of integrated analysis on heterogeneous data. Other R packages for DNA methylation analysis have community supports; however, they require external data preprocessing, read mapping and methylation calling to generate base-resolution DNA methylation data.
EpiMOLAS is unique among the currently available WGBS analysis platforms and tools in many aspects. Most workflows and tools provide graphical results, but they are limited to specific types of analyses. In EpiMOLAS_web, we designed various modules for quantitative analyses as well as a keyword-based query on the gene annotations. Taking advantage of web servers, users can explore their data in various ways and save the gene lists of each analysis with tracking logs. By utilizing BioGRID protein interaction data, we can further study the association of the genes discovered in a methylome analysis and their protein interaction network in an interactome analysis.
EpiMOLAS adopts differentially methylated genes (DMGs) instead of differentially methylated regions or cytosines (DMRs or DMCs, respectively). On the basis of DMGs, this approach would flatten the impact of DMRs into broad-scale regional signals and make it a complementary view of studying aberrant DNA methylation regions with specific genomic features base by base. Moreover, we use a straightforward approach to assess the DMGs calculated based on pairwise comparison or on a preset background subtraction. From a macro perspective, we provide a “gene-centric” approach to study the genome-wide DNA methylation changes with their potential biological functions via downstream gene set analysis.
Conclusions
To the best of our knowledge, EpiMOLAS is the first web-based framework that adopts a two-phase approach to process WGBS raw reads and provides versatile downstream analysis, annotation, and visualization, enabling users to explore their data and obtain useful information. Docker containerization technology applied in the streamlined DNA methylation profiling workflow is not only rapid for deployment but up-scalable by increasing the number of containers running in a cloud computing environment, thereby meeting the needs of various scales of experimental design. EpiMOLAS helps users deal with their WGBS data and furthermore, alleviates the burdens of conducting reproducible analyses of publicly available data.
Availability and requirements
Project name: EpiMOLAS.
Project home page:
http://symbiosis.iis.sinica.edu.tw/epimolas/ (EpiMOLAS_web).
https://hub.docker.com/r/lsbnb/docmethyl/ (DocMethyl).
https://github.com/markchiang/EPI-MOLAS/ (EpiMolas.jar).
Operating system(s): Docker containers and web application are platform-independent.
Programming language: Java, Python and R.
Other requirement: Docker installation for DocMethyl use.
License: Galaxy source code is licensed under the Academic Free License version 3.0.
Any restrictions to use by non-academics: No.
Availability of data and materials
Not applicable.
Abbreviations
- BS-seq:
-
Bisulfite sequencing
- DMC:
-
Differentially methylated cytosine
- DMG:
-
Differentially methylated gene
- DMR:
-
Differentially methylated region
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- MBD-seq:
-
Methyl-CpG-binding domain (MBD) proteins with sequencing
- MeDIP-seq:
-
Methylated DNA immunoprecipitation with sequencing
- PPIN:
-
Protein-protein interaction network
- RRBS:
-
Reduced representation bisulfite sequencing
- WGBS:
-
Whole-genome bisulfite sequencing
References
Li E, Zhang Y. DNA methylation in mammals. Cold Spring Harb Perspect Biol. 2014;6:a019133.
To TK, Saze H, Kakutani T. DNA methylation within transcribed regions. Plant Physiol. 2015;168:1219–25.
Serre D, Lee BH, Ting AH. MBD-isolated genome sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010;38(2):391–9.
Taiwo O, Wilson GA, Morris T, Seisenberger S, Reik W, Pearce D, Beck S, Butcher LM. Methylome analysis using MeDIP-seq with low DNA concentrations. Nat Protoc. 2012;7(4):617–36.
Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33(18):5868–77.
Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452(7184):215–9.
Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, Van Djik S, Muhlhausler B, Stirzaker C, Clark SJ. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17(1):208.
Lienhard M, Grimm C, Morkel M, Herwig R, Chavez L. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics. 2014;30(2):284–6.
Huang J, Renault V, Sengenes J, Touleimat N, Michel S, Lathrop M, Tost J. MeQA: a pipeline for MeDIP-seq data quality assessment and analysis. Bioinformatics. 2012;28(4):587–8.
Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26(7):779–85.
Kubsad D, Nilsson EE, King SE, Sadler-Riggleman I, Beck D, Skinner MK. Assessment of glyphosate induced epigenetic Transgenerational inheritance of pathologies and sperm Epimutations: generational toxicology. Sci Rep. 2019;9(1):6372.
Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89(5):1827–31.
Adusumalli S, Mohd Omar MF, Soong R, Benoukraf T. Methodological aspects of whole-genome bisulfite sequencing analysis. Brief Bioinform. 2015;16:369–79.
Tsuji J, Weng Z. Evaluation of preprocessing, mapping and postprocessing algorithms for analyzing whole genome bisulfite sequencing data. Brief Bioinform. 2016;17:938–52.
Yong WS, Hsu FM, Chen PY. Profiling genome-wide DNA methylation. Epigenetics Chromatin. 2016;9:26.
Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Eberhard C, et al. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–W10.
Andrews SR. FastQC: a quality control tool for high throughput sequence data; 2010.
Krueger F. Trim Galore: A wrapper around Cutadapt and FastQC to consistently apply adapter and quality trimming to FastQ files; 2012.
Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for bisulfite-Seq applications. Bioinformatics. 2011;27:1571–2.
Su SY, Chen SH, Lu IH, Chiang YS, Wang YB, Chen PY, Lin CY. TEA: the epigenome platform for Arabidopsis methylome study. BMC Genomics. 2016;17:1027.
Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Giron CG, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–61 Accessed 27 Feb. 2019.
Down TA, Piipari M, Hubbard TJ. Dalliance: interactive genome viewing on the web. Bioinformatics. 2011;27:889–90.
Fernandez NF, Gundersen GW, Rahman A, Grimes ML, Rikova K, Hornbeck P, Ma'ayan A. Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data. Sci Data. 2017;4:170151.
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9 Accessed 27 Feb. 2019.
Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30 Accessed 27 Feb. 2019.
Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, O'Donnell L, Oster S, Theesfeld C, Sellam A, et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45:D369–79 Accessed 27 Feb. 2019.
Franz M, Lopes CT, Huck G, Dong Y, Sumer O, Bader GD. Cytoscape.Js: a graph theory library for visualisation and analysis. Bioinformatics. 2016;32:309–11.
Kretzmer H, Otto C, Hoffmann S. BAT: Bisulfite Analysis Toolkit. F1000Res. 2017;6:1490.
JENCODE WGBS pipeline is available at https://www.encodeproject.org/data-standards/wgbs/. Accessed 27 Feb. 2019.
Bhardwaj V, Heyne S, Sikora K, Rabbani L, Rauer M, Kilpert F, Richter AS, Ryan DP, Manke T. snakePipes enable flexible, scalable and integrative epigenomic analysis. bioRxiv. 2018, 407312. https://doi.org/10.1101/407312.
Ewels PA, Peltzer A, Fillinger S, Alneberg JA, Patel H, Wilm A, Garcia MU, Di Tommaso P, Nahnsen S. nf-core: Community curated bioinformatics pipelines. bioRxiv. 2019; p. 610741. https://doi.org/10.1101/610741.
Cavalcante RG, Patil S, Park Y, Rozek LS, Sartor MA. Integrating DNA methylation and Hydroxymethylation data with the mint pipeline. Cancer Res. 2017;77:e27–30.
Kishore K, de Pretis S, Lister R, Morelli MJ, Bianchi V, Amati B, Ecker JR. Pelizzola M: methylPipe and compEpiTools: a suite of R packages for the integrative analysis of epigenomics data. BMC Bioinformatics. 2015;16:313.
Park Y, Figueroa ME, Rozek LS, Sartor MA. MethylSig: a whole genome DNA methylation analysis pipeline. Bioinformatics. 2014;30:2414–22.
Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A. Mason CE: methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87.
Acknowledgments
The authors thank anonymous reviewers for providing comments and suggestions that helped us improve quality and clarity of the manuscript.
About this supplement
This article has been published as part of BMC Genomics, Volume 21 Supplement 3, 2020: 18th International Conference on Bioinformatics. The full contents of the supplement are available at https://bmcgenomics.biomedcentral.com/articles/supplements/volume-21-supplement-3.
Funding
The authors thank Ministry of Science and Technology (MOST), Taiwan, for financially supporting this research and publication through MOST108–2314-B-001-002, MOST107–2321-B-002-057, MOST108–2321-B-038-003, MOST 108–2321-B-037-001 to CYL, and flagship program from Institute of Information Science, Academia Sinica, Taiwan to CYL and JMH. The funding body initially identified the general research field that aligned with their funded scopes. Individuals working for the funding body played the roles in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript as mentioned in the Authors’ contributions.
Author information
Authors and Affiliations
Contributions
SYS, PYC, JMH, SHC and CYL conceived and designed the project. SYS, IHL and WCC1 constructed the web server and integrated the data analysis workflows and visualization. SYS and WCC2 conducted the implementation of Galaxy Docker Image. SYS, SHC and CYL wrote the manuscript. SYS, SHC and CYL participated in discussions and conceptualization as well as revising the draft. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
This supplementary file provides the description and usage of DocMethyl and EpiMOLAS_web, including the installation steps on how to execute the workflow in DocMethyl and the usage guides on analyzing WGBS data through the modules in EpiMOLAS_web.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Su, SY., Lu, IH., Cheng, WC. et al. EpiMOLAS: an intuitive web-based framework for genome-wide DNA methylation analysis. BMC Genomics 21 (Suppl 3), 163 (2020). https://doi.org/10.1186/s12864-019-6404-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-6404-8