Abstract
Background
We previously conducted a genome-wide association study (GWAS) strategy for milk fatty acids in Chinese Holstein, and identified 83 genome-wide significant single nucleotide polymorphisms (SNPs) and 314 suggestive significant SNPs. Among them, two SNPs, BTB-01077939 and BTA-11275-no-rs associated with C10:0, C12:0, and C14 index (P = 0.000014 ~ 0.000024), were within and close to (0.85 Mb) protein kinase, cGMP-dependent, type І (PRKG1) gene on BTA26, respectively. PRKG1 gene plays a key role in lipolysis to release fatty acids and glycerol through the hydrolysis of triacyglycerol in adipocytes. We herein considered it as a promising candidate for milk fatty acids. The purpose of this study was to investigate whether PRKG1 had effects on milk fatty acids.
Results
By direct sequencing the PCR products of pooled DNA, we identified a total of six SNPs, including one in 5′ flanking region, four in 3′ untranslated region (UTR), and one in 3′ flanking region. The single-locus association analysis was carried out, and showed that the six SNPs mainly had significant associations with C6:0, C8:0 and C17:1 (P < 0.0001 ~ 0.0035). In addition, we observed a haplotype block formed by g.6903810G > A and g.6904047G > T with Haploview 4.1, and it was strongly associated with C8:0, C10:0, C16:1, C17:1, C20:0 and C16 index (P = < 0.0001 ~ 0.0123). The SNP, g.8344262A > T, was predicted to alter the binding site (BS) of transcription factor (TF) GAGA box with Genomatix software, and the subsequent luciferase assay verified that it really changed the transcriptional activity of PRKG1 gene (P = 0.0009).
Conclusion
In conclusion, to our best of knowledge, we are the first who identified the significant effects of PRKG1 on milk fatty acids in dairy cattle.
Similar content being viewed by others
Background
Dairy products are well known for vital nutrients providing high quality protein and energy in human diet [1,2,3]. The most important economic traits of milk production in dairy cattle include milk yield, fat and protein yield, and fat and protein percentage [4]. Milk fat contains a lot of fatty acids composed of saturated fatty acid (SFA) and unsaturated fatty acid (UFA), and it determines the physiological and sensory properties of the milk [5]. SFA increases the risk of cardiovascular diseases, while UFA decreases the risk [6,7,8,9,10,11]. For example, C12:0, C14:0 and C16:0 have adverse effects on lower-density lipoprotein cholesterol, that is a risk factor for cardiovascular diseases [10], and substituting n-6 and n-3 polyunsaturated fatty acids for SFAs decreases cardiovascular diseases morbidity and mortality [11]. Fatty acids are regulated by a huge network of genes encoding transcription and translational regulators in living organisms [12], and the heritability of SFA (0.14 ~ 0.33) and UFA (0.08 ~ 0.29) have been reported [13,14,15,16,17].
In dairy cattle, some promising candidate genes and QTL regions for milk fatty acids have been identified in previous Genome-wide association studies (GWASs), such as fatty acid synthase (FASN; on BTA19), diacylglycerol O-acyltransferase 1 (DGAT1; BTA14), stearoyl-CoA desaturase (SCD; on BTA26), 22,833,168 bp to 26,284,743 bp on BTA9 and 1,588,879 bp to 2,764,862 bp on BTA14 [18,19,20,21,22,23]. Our previous GWAS [23] discovered 83 genome-wide significant single nucleotide polymorphisms (SNPs) and 314 suggestive significant SNPs associated with milk fatty acids in Chinese Holstein cows, in which, one SNP, BTB-01077939 for C10:0 (P = 0.000014) and C14 index (P = 0.000014), was located within the protein kinase, cGMP-dependent, type І (PRKG1) gene, and the other SNP, BTA-11275-no-rs for C12:0 (P = 0.000024), was close to the PRKG1 gene with a distance of 0.85 Mb. In addition, Li et al. reported that a QTL region (1.00 Mbp ~ 27.94 Mbp) on BTA26 is significantly associated with milk fatty acids by performing a joint GWAS based on Chinese and Danish Holstein populations [24]. PRKG1 gene is located on BTA26 (6,901,760 ~ 8,343,635 bp) spanning about 1442 kb and includes 20 exons. It regulates the lipolysis in adipocytes to release fatty acids and glycerol by the hydrolysis of triacyglycerol. It was reported that PRKG1 gene was involved in cGMP-PKG signaling pathway to inhibit rat brown adipocyte proliferation [25]. To date, no study has reported the associations of PRKG1 gene with milk fatty acids in dairy cattle. Hence, the objective of this study was to detect whether the PRKG1 gene had effects on milk fatty acids. We herein searched potential SNPs in PRKG1 gene, and examined associations of the identified SNPs with 24 traits in Chinese Holstein cows. Further, we verified the impact of one regulatory SNP on transcriptional activity of PRKG1 with dual-luciferase assay.
Results
SNPs identification
By screening the entire coding region and 5′ and 3′ flanking regions, we identified six SNPs (Table 1) in PRKG1 gene, including g.8344262A > T in 5′ flanking region, g.6904047G > T, g.6903810G > A, g.6903365C > A and g.6902878 T > G in 3′ untranslated region (UTR), and g.6901713 T > G in 3′ flanking region. The genotypic and allele frequencies of the six SNPs in PRKG1 gene were shown in Table 1.
Associations between SNPs/haplotype blocks and 24 milk fatty acids
Genetic associations (significant associations with P < 0.0001 ~ 0.0035) of six SNPs were detected for 24 milk fatty acids, and the results were shown in Table 2. The g.8344262A > T was significantly associated with C10:0, C18:1cis-9 and total index. The g.6904047G > T was significantly associated with C17:1 and C17 index. The g.6903810G > A was significantly associated with C8:0, C20:0 and total index. The g.6903365C > A was significantly associated with C6:0, C8:0 and C20:0. The g.6902878 T > G was significantly associated with C6:0, C8:0 and C17:1. The g.6901713 T > G was significantly associated with C6:0, C8:0, C10:0, C17:1, C14 index and C17 index. In addition, as shown in Table 3, the significant dominant (a), additive (d) and allele substitution (α) effects of these six SNPs for C6:0, C8:0, C10:0, C12:0, C14:0, C14:1, C16:0, C16:1, C17:1, C18:1cis-9, C18 index, C20:0, C14 index, C16 index, C17 index, SFA, UFA, SFA/UFA and total index were presented in Table 3 (P < 0.05).
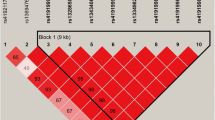
With the Haploview 4.1, we found a haplotype block (Fig. 1) formed by two SNPs, g.6904047G > T and g.6903810G > A. The haplotype block included haplotypes GG, GT and AG with their frequencies of 67.70, 20.00 and 12.30%, respectively. The haplotype-based association analysis (Table 4) showed that the haplotype block had significant associations with C8:0, C10:0, C16:1, C17:1, C20:0 and C16 index (P < 0.0001 ~ 0.0123).
Further, by estimating the LD among the SNPs of PRKG1 and SCD genes with a distance of 12.79 Mbp, we did not found haplotype block between the two genes (Fig. 2), indicating that the significant effects of PRKG1 gene on milk fatty acids were not induced by LD with SCD gene.
Change of transcriptional activity caused by g.8344262A > T
We predicted the change of TFBS caused by the SNP in the 5′ flanking region of PRKG1 gene using Genomatix software, and found that the allele T of g.8344262A > T created a TFBS for GAGA-Box (GAGA).
To detect whether g.8344262A > T changed the transcriptional activity of PRKG1, we synthesized two constructs with A and T of g.8344262A > T, respectively (Fig. 3a). We measured the luciferases activities of firefly and renilla, and showed the results in Fig. 3b. Luciferase activities of the two constructs were significantly higher than that of the blank control (P ≤ 0.0005) and empty vector (PGL4.14; P ≤ 0.0006), suggesting that g.8344262A > T might have the transcriptional activity. The allele T of g.8344262A > T had significantly lower luciferase activity than the allele A (P = 0.0009), implying that g.8344262A > T could alter the transcriptional activity of PRKG1 gene.
Dual-luciferase assay. a Sketches of recombinant plasmids with g.8344262A > T in the 5′ flanking region of PRKG1 gene. The nucleotides in red highlight referred to the SNP. b Luciferase assay analysis of the recombinant plasmids in HEK293 cells. Blank: Blank cells. PGL4.14: Empty vector. A and T: Plasmids of g.8344262A > T. **: P < 0.0005
Discussion
In our initial GWAS [23], PRKG1 was considered as one of the promising candidate gene on milk fatty acids in Chinese Holstein. It was also reported that PRKG1 gene was associated with fatty acid composition in swine [26], and the PRKG1 knockout mice had lower triglyceride stores in the brown adipose tissue [27]. Here, we first verified that PRKG1 gene had significant effects on medium-chain saturated fatty acids in dairy cattle, especially C8:0.
SCD gene was also a promising candidate gene on BTA26 for milk fatty acids [23], and its effects had been confirmed [28]. In a joint GWAS based on Chinese and Danish Holstein populations, Li et al. also identified a significant QTL region for milk fatty acids (10.00 ~ 27.94 Mbp on BTA26), which included SCD and PRKG1 [24], and the SCD was in downstream of PRKG1 with a distance of 12.79 Mbp. In this study, it was shown that no LD among the SNPs of PRKG1 and SCD was observed, indicating that the effects of PRKG1 on milk fatty acid traits were independent from SCD.
From the KEGG database, we found that PRKG1 was involved in the cGMP-PKG signaling pathway (ko04022) and regulation of lipolysis in adipocytes (ko04923). In rat, cGMP signaling inhibited brown adipocyte proliferation and thereby promoted brown adipocyte differentiation [25]. The brown adipose tissue from PRKG1 knockout mice decreased triglyceride stores, suggesting an increase in the ratio of pre-adipocytes to adipocytes and fewer fully differentiated brown adipocytes [27]. In swine, the RNA-Seq analysis identified that the PRKG1 gene was the differentially-expressed in muscle between high and low groups for fatty acid composition traits [29]. Considering the significant effect of PRKG1 on milk fatty acid in the present study, we deduced that the gene might have important regulatory function for milk fatty acid metabolism in dairy cattle.
Gene expression is commonly controlled by TFs that are bound to specific sequence elements and the regular regions of the genome [30, 31]. TFBSs are the biding sites (BS) targeted by a DNA binding protein [32], and its disruption is associated with phenotypic diversity [33, 34]. In this study, the allele T of g.8344262A > T was predicted to create a BS for TF GAGA, and it significantly lowered the transcriptional activity of PRKG1. GAGA is a drosophila transcription factor involved in many nuclear activities, and can enhance transcription by stabilizing pre-initiation complex and promoting re-initiation [35]. Interactions of GAGA-binding proteins with the GAGA of the V1bR promoter activate V1bR gene expression, and provide a potential mechanism for physiological regulation of V1bR transcription [36]. The significant associations of g.8344262A > T with milk fatty acids, and its impact on transcriptional activity of PRKG1 gene, suggested that g.8344262A > T might be a potentially causal mutation regulating the PRKG1 expression by changing the BS for the TF GAGA to affect the formation of milk fatty acids in dairy cattle.
Conclusions
According to our previous GWAS, we considered PRKG1 gene as a promising candidate for milk fatty acids in dairy cattle. In the present study, we further confirmed the effects of PRKG1 on milk fatty acids, and showed that the gene mainly impact on medium-chain saturated fatty acid traits. In addition, we revealed that g.8344262A > T might be a potentially causal mutation altering the transcriptional activity due to the change of a BS for TF GAGA. Our findings might be helpful for the marker-assisted selection in dairy cattle.
Methods
Animals and measures of milk fatty acids
We used 1065 Chinese Holstein cows from 44 sire families with an average of 24 daughters per sire for the association analyses in this study. The cows were from 23 dairy farms of Sanyuanlvhe Dairy Farming Center (Beijing, China), a leading dairy company in China, where the standard performance testing for dairy herd improvement (DHI) has been regularly conducted since 1999, and all the cows were fed with the same regular total mixed ration (TMR) composed of concentrated feed and coarse fodder across all subordinate farms. From November to December of 2014, we collected 50 mL milk samples for each cow during 1 ~ 240 days of the first lactation.
Then, we used 2 mL milk samples to measure 16 milk fatty acids (C6:0, C8:0, C10:0, C11:0, C12:0, C13:0, C14:0, C15:0, C16:0, C17:0, C18:0, C20:0, C14:1, C16:1, C17:1 and C18:1cis-9) with the gas chromatography described in the previous GWAS [23] in Beijing Dairy Cattle Center (www.bdcc.com.cn). In addition, we calculated C14 index, C16 index, C17 index, C18 index and total index based on the formulas: \( \frac{\mathrm{cis}-9\ \mathrm{unsaturated}}{\mathrm{cis}-9\ \mathrm{unsaturated}+\mathrm{saturated}}\ast 100 \) [37], and summarized SFA, UFA, SFA/UFA.
SNP identification and genotyping
We extracted semen DNAs of 44 Holstein bulls who were sires of the above-mentioned cows using the salt-out procedure, and extracted blood DNAs of the 1065 Chinese Holstein cows with the TIANamp Blood DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. We measured the quantity and quality of extracted DNA using a NanoDrop™ ND-2000 Spectrophotometer (Thermo Scientific, Hudson, DE, USA) and 2% agarose gel electrophoresis, respectively.
Based on the genomic sequence of the bovine PRKG1 (Gene ID: 282004), we designed 33 pairs of primers (Additional file 1: Table S1) corresponding the entire exons and 3000 bp of 5′ and 3′ flanking regions using the Primer 3.0 (http://primer3.wi.mit.edu/). The primers were synthesized in Beijing Genomics Institute (Beijing Genomic Institute, Beijing, China). We diluted the genomic DNA of each bull into the concentration of 50 ng/μL, and randomly pooled them into two equal pools (each pool included 22 sire DNAs). The final reaction volume of PCR included 2 μL genomic DNA, 1.25 μL each primer (10 mM), 12.5 μL Premix TaqTM (Takara, Dalian, China) and 8 μL DNase/RNase-Free Deionized Water (Tiangen, Beijing, China).The PCR amplifications for the pooled DNAs were performed, and the procedures were as follows: initial denaturation at 94 °C for 5 min, followed by 35 cycles of 30s at 94 °C, annealing at 60 °C for 30s, extension at 72 °C for 30s, and a final extension at 72 °C for 7 min. Then, we bidirectionally sequenced the PCR products by ABI3730xl DNA analyzer (Applied Biosystems, CA, USA), and aligned them with the bovine reference sequences (UMD 3.1.1) using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to search the potential SNPs.
For the SNPs identified in PRKG1 gene, we used the matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS, Sequenom MassARRAY, Bioyong Technologies Inc. HK) to perform the genotyping for the 1065 Chinese Holstein cows.
Prediction of the transcription factor binding site (TFBS)
We used Genomatix software suite v3.9 (http://www.genomatix.de/cgi-bin/welcome/welcome.pl?s=d1b5c9a9015b02bb3b1a806f9c03293f) to predict the change of TFBS caused by the SNP g.8344262A > T in 5′ flanking region of PRKG1 gene. In the prediction, we input 30 bp of up/down-stream sequences of g.8344262A > T, namely AGTTTAATATTTATGAAATGTCTCTCTCTC[A]CACACACACACACACACACTCACACGCACA and AGTTTAATATTTATGAAATGTCTCTCTCTC[T]CACACACACACACACACACTCACACGCACA, to research the different transcription factor (TF) bound for allele A and T.
Recombinant plasmid construction and luciferase assay
In this study, we used the luciferase assay to verify whether the SNP g.8344262A > T changed the transcriptional activity of PRKG1 gene. We synthesized (Genewiz, Suzhou, China) two fragments (Fig. 3a), A and T of g.8344262A > T, including NheI and HindIII restriction sites at the 5′ to 3′ termini, respectively, and cloned them into the pGL4.14 Luciferase Assay Vector (Promega, Madison, USA). We sequenced these two plasmid constructs to confirm the integrity of the insertions. Then, we purified all the plasmids using the Endo-free Plasmid DNA Mini Kit II (OMEGA, omega bio-tek, Norcross, Georgia, USA).
We cultured Human Embryonic Kidney (HEK)-293 T cells in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Life Technologies, USA) containing 10% heat-inactivated fetal bovine serum (FBS; Gibco) at 5% CO2 and 37 °C. We seeded approximately 2 × 105 cells per well in the 24-well plates, and then used Lipofectamine 2000 (Invitrogen, CA, USA) to transfect the cells according to the manufacturer’s protocol. We transfected 500 ng constructed plasmid DNA along with 10 ng of pRL-TK renilla luciferase reporter vector (Promega) into each well. The experiments were conducted in three replicates for each construct.
We harvested the cells about 48 h after transfection, and measured the activities of firefly and renilla luciferases using a Dual-Luciferase Reporter Assay System (Promega, Madison, USA) on a Modulus microplate multimode reader (Turner Biosystems, CA, USA). We used the average statistics of three replicates as the normalized luciferase data (firefly/renilla).
Estimation of the linkage disequilibrium (LD)
We estimated the LD among the SNPs identified in PRKG1 gene in this study using Haploview 4.1 (Broad Institute of MIT and Harvard, Cambridge, MA, USA), and identified the haplotype block. In the process, 95% confidence bounds on D’ were generated and each comparison was called “strong LD”, “inconclusive” or “strong recombination”. If 95% of informative (i.e. non-inconclusive) comparisons were “strong LD”, a block would be created [38]. In addition, we used the r2 to represent the correlation coefficient between two loci.
We also performed LD analysis between 8 SNPs in PRKG1 gene and 24 SNPs in SCD gene to detect whether the significant effects of PRKG1 on milk fatty acids were caused by SCD. As the significant SNPs of the two genes identified in two previous studies (Additional file 2: Table S2) [23, 28] were not genotyped in this study, we herein used the database of 1000 Bull Genomes Project [39] to estimate the LD between PRKG1 and SCD. The database of 1000 Bull Genomes Project, including 1575 individuals involved in 48 breeds (Additional file 3: Table S3), was different from our population, while its worldwide correlation implied that the result could be as the indirect support for this study.
Association analysis
We analyzed the associations between each SNP/haplotype block and 24 milk fatty acids using SAS9.2 software (SAS INSTITUTE Inc., Cary, NC, USA) with the following mixed animal model:
For each fatty acid trait, Y is the phenotypic value; mu is the overall mean; herd is the fixed effect of farm; lactation stage is the fixed effect of stage of lactation; M is the fixed effect of calving month; b is the regression coefficient of covariate M; G is the fixed effect corresponding to the genotype or haplotype combination; A is the random polygenic effect that is distributed as N (0, A \( {\sigma}_a^2 \)), in which, the numerator relationship matrix (A-matrix) was constructed by using Fortran95 code. Pedigree information of the genotyped animals was traced back for three generations. As a result, the total number of animals included in the analysis reaches 3335. In addition, e is the random residual, distributed as N (0, I \( {\sigma}_e^2 \)), with identity matrix I and residual error variance \( {\sigma}_e^2 \). The Bonferroni correction for multiple testing was performed based on the number of SNPs. The significant levels of the single SNPs after correction for multiple testing at P < 0.05 and P < 0.01 were 0.0083 and 0.0017, respectively. In addition, we calculated the additive effect (a), dominant effect (d), and substitution effect (α) using the following formulas [40]: \( \alpha =\frac{AA- BB}{2},d= AB-\frac{AA+ BB}{2}, and\ \alpha =\alpha +d\left(q-p\right) \). In which, AA, AB and BB are the least square means of fatty acid traits corresponding to the genotypes, and p and q are the frequencies of A and B, respectively. Here, the P values for significant effects were P < 0.05 and P < 0.01.
Availability of data and materials
All relevant data are available within the article and its additional files.
Abbreviations
- a:
-
Additive effect
- BS:
-
Binding site
- d:
-
Dominant effect
- DHI:
-
Dairy herd improvement
- GWAS:
-
Genome-wide association study
- LD:
-
Linkage disequilibrium
- PRKG1 :
-
Protein kinase, cGMP-dependent, type І
- SFA:
-
Saturated fatty acids
- SNP:
-
Single nucleotide polymorphism
- TF:
-
Transcription factor
- TFBS:
-
Transcription factor binding site
- UFA:
-
Unsaturated fatty acids
- UTR:
-
Untranslated region
- α:
-
Substitution effect
References
Designing foods: animal product options in the marketplace. Washington (DC); 1988. Copyright © 1988 by the National Academy of Sciences.
Lindsay H. Allen. Animal source foods to improve micronutrient nutrition and human function in developing countries. Proceedings of a conference. June 24-26, 2002. Washington, DC, USA. J Nutr. 2003;133(11 Suppl 2):3875S–4061S.
Haug A, Hostmark AT, Harstad OM. Bovine milk in human nutrition--a review. Lipids Health Dis. 2007;6:25.
Spelman RJ, Coppieters W, Karim L, van Arendonk JA, Bovenhuis H. Quantitative trait loci analysis for five milk production traits on chromosome six in the Dutch Holstein-Friesian population. Genetics. 1996;144(4):1799–808.
Illingworth D. Handbook of milkfat fractionation technology and applications : by Kerry E. Kaylegian and Robert C. Lindsay, AOCS press, 1995. $150.00 (xxiv + 662 pages) ISBN 0 935315 57 8. Trends Food Sci Technol. 1996;7(1):32–3.
Briggs MA, Petersen KS, Kris-Etherton PM. Saturated fatty acids and cardiovascular disease: replacements for saturated fat to reduce cardiovascular risk. Healthcare. 2017;5(2).
Mensink RP, Zock PL, Kester ADM, Katan MB. Effects of dietary fatty acids and carbohydrates on the ratio of serum total to HDL cholesterol and on serum lipids and apolipoproteins: a meta-analysis of 60 controlled trials. Am J Clin Nutr. 2003;77(5):1146–55.
Kris-Etherton PM, Pearson TA, Wan Y, Hargrove RL, Moriarty K, Fishell V, Etherton T. High-monounsaturated fatty acid diets lower both plasma cholesterol and triacylglycerol concentrations. Am J Clin Nutr. 1999;70(6):1009–15.
Kromhout D, Menotti A, Kesteloot H, Sans S. Prevention of coronary heart disease by diet and lifestyle - evidence from prospective cross-cultural, cohort, and intervention studies. Circulation. 2002;105(7):893–8.
Fernandez ML, West KL. Mechanisms by which dietary fatty acids modulate plasma lipids. J Nutr. 2005;135(9):2075–8.
Kris-Etherton PM, Fleming JA. Emerging nutrition science on fatty acids and cardiovascular disease: nutritionists' perspectives. Adv Nutr. 2015;6(3):326S–37S.
Bionaz M, Loor JJ. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics. 2008;9:366.
Stoop WM, Schennink A, Visker MH, Mullaart E, van Arendonk JA, Bovenhuis H. Genome-wide scan for bovine milk-fat composition. I. Quantitative trait loci for short- and medium-chain fatty acids. J Dairy Sci. 2009;92(9):4664–75.
Schennink A, Stoop WM, Visker MH, van der Poel JJ, Bovenhuis H, van Arendonk JA. Short communication: genome-wide scan for bovine milk-fat composition. II. Quantitative trait loci for long-chain fatty acids. J Dairy Sci. 2009;92(9):4676–82.
Petrini J, Iung LH, Rodriguez MA, Salvian M, Pertille F, Rovadoscki GA, Cassoli LD, Coutinho LL, Machado PF, Wiggans GR, et al. Genetic parameters for milk fatty acids, milk yield and quality traits of a Holstein cattle population reared under tropical conditions. J Anim Breed Genet = Z Tierzuecht Zuechtungsbiol. 2016;133(5):384–95.
Krag K, Poulsen NA, Larsen MK, Larsen LB, Janss LL, Buitenhuis B. Genetic parameters for milk fatty acids in Danish Holstein cattle based on SNP markers using a Bayesian approach. BMC Genet. 2013;14:79.
Narayana SG, Schenkel FS, Fleming A, Koeck A, Malchiodi F, Jamrozik J, Johnston J, Sargolzaei M, Miglior F. Genetic analysis of groups of mid-infrared predicted fatty acids in milk. J Dairy Sci. 2017;100(6):4731–44.
Olsen HG, Knutsen TM, Kohler A, Svendsen M, Gidskehaug L, Grove H, Nome T, Sodeland M, Sundsaasen KK, Kent MP, et al. Genome-wide association mapping for milk fat composition and fine mapping of a QTL for de novo synthesis of milk fatty acids on bovine chromosome 13. Genet Sel Evol. 2017;49(1):20.
Buitenhuis B, Janss LL, Poulsen NA, Larsen LB, Larsen MK, Sorensen P. Genome-wide association and biological pathway analysis for milk-fat composition in Danish Holstein and Danish Jersey cattle. BMC Genomics. 2014;15:1112.
Li X, Buitenhuis AJ, Lund MS, Li C, Sun D, Zhang Q, Poulsen NA, Su G. Joint genome-wide association study for milk fatty acid traits in Chinese and Danish Holstein populations. J Dairy Sci. 2015;98(11):8152–63.
Palombo V, Milanesi M, Sgorlon S, Capomaccio S, Mele M, Nicolazzi E, Ajmone-Marsan P, Pilla F, Stefanon B, D'Andrea M. Genome-wide association study of milk fatty acid composition in Italian Simmental and Italian Holstein cows using single nucleotide polymorphism arrays. J Dairy Sci. 2018;101(12):11004-19.
Knutsen TM, Olsen HG, Tafintseva V, Svendsen M, Kohler A, Kent MP, Lien S. Unravelling genetic variation underlying de novo-synthesis of bovine milk fatty acids. Sci Rep. 2018;8(1):2179.
Li C, Sun DX, Zhang SL, Wang S, Wu XP, Zhang Q, Liu L, Li YH, Qiao L. Genome wide association study identifies 20 novel promising genes associated with Milk fatty acid traits in Chinese Holstein. PLoS One. 2014;9(5):e96186.
Li X, Buitenhuis AJ, Lund MS, Li C, Sun D, Zhang Q, Poulsen NA, Su G. Joint genome-wide association study for milk fatty acid traits in Chinese and Danish Holstein populations. J Dairy Sci. 2015;98(11):8152–63.
Nisoli E, Clementi E, Tonello C, Sciorati C, Briscini L, Carruba MO. Effects of nitric oxide on proliferation and differentiation of rat brown adipocytes in primary cultures. Br J Pharmacol. 1998;125(4):888–94.
Revilla M, Puig-Oliveras A, Castello A, Crespo-Piazuelo D, Paludo E, Fernandez AI, Ballester M, Folch JM. A global analysis of CNVs in swine using whole genome sequence data and association analysis with fatty acid composition and growth traits. PLoS One. 2017;12(5):e0177014.
Amieux PS, McKnight GS. Cyclic nucleotides converge on brown adipose tissue differentiation. Sci Signal. 2010;3(104):pe2.
Li C, Sun D, Zhang S, Liu L, Alim MA, Zhang Q. A post-GWAS confirming the SCD gene associated with milk medium- and long-chain unsaturated fatty acids in Chinese Holstein population. Anim Genet. 2016;47(4):483–90.
Puig-Oliveras A, Ramayo-Caldas Y, Corominas J, Estelle J, Perez-Montarelo D. Differences in muscle Transcriptome among pigs phenotypically extreme for fatty acid composition. PLoS One. 2014;9(6):e99720.
Fuda FS, Karandikar NJ, Chen W. Significant CD5 expression on normal stage 3 hematogones and mature B lymphocytes in bone marrow. Am J Clin Pathol. 2009;132(5):733–7.
Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108(4):439–51.
Yu CP, Li WH. Predicting transcription factor binding sites and their cognate transcription factors using gene expression data. Methods Mol Biol. 2017;1629:271–82.
Swinnen G, Goossens A, Pauwels L. Lessons from domestication: targeting Cis-regulatory elements for crop improvement. Trends Plant Sci. 2016;21(6):506–15.
Deplancke B, Alpern D, Gardeux V. The genetics of transcription factor DNA binding variation. Cell. 2016;166(3):538–54.
Vaquero A, Blanch M, Espinas ML, Bernues J. Activation properties of GAGA transcription factor. Biochim Biophys Acta. 2008;1779(5):312–7.
Volpi S, Rabadan-Diehl C, Cawley N, Aguilera G. Transcriptional regulation of the pituitary vasopressin V1b receptor involves a GAGA-binding protein. J Biol Chem. 2002;277(31):27829–38.
Kelsey JA, Corl BA, Collier RJ, Bauman DE. The effect of breed, parity, and stage of lactation on conjugated linoleic acid (CLA) in milk fat from dairy cows. J Dairy Sci. 2003;86(8):2588–97.
Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–9.
Daetwyler HD, Capitan A, Pausch H, Stothard P, van Binsbergen R, Brondum RF, Liao X, Djari A, Rodriguez SC, Grohs C, et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat Genet. 2014;46(8):858–65.
Falconer DS, Mackay TFC: Introduction to quantitative genetics. 1996: Ed. 4 xv + 464 pp.
Acknowledgements
We appreciate Beijing Dairy Cattle Center, Beijing Municipal Bureau of Agriculture, and Beijing Sanyuanlvhe Dairy Farming Center, for providing the milk, semen, and blood samples of Chinese Holstein.
Funding
This work was financially supported by the Beijing Dairy Industry Innovation Team (BAIC06–2018/2019), National Natural Science Foundation of China (31872330, 31802041), Beijing Science and Technology Program (D171100002417001), National Science and Technology Programs of China (2013AA102504), earmarked fund for Modern Agro-industry Technology Research System (CARS-36), and the Program for Changjiang Scholar and Innovation Research Team in University (IRT_15R62). These funding bodies had no roles in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
DS conceived and designed the experiments, LL, YY and ZM prepared the milk, blood and semen samples, LS extracted the DNA for SNP identification and genotyping with the help of XL, XL measured the milk fatty acids, LS analyzed the data, and the manuscript was prepared by LS, BH and DS. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All protocols for collection of the samples of experimental individuals and phenotypic observations were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at China Agricultural University (Permit Number: DK996). Milk, blood and semen samples were collected specifically for this study following standard procedures with the full agreement of the Beijing Sanyuanlvhe Dairy Farming Center who owned the Holstein cows and bulls, respectively.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. PCR primer information of PRKG1 gene. (XLSX 12 kb)
Additional file 2:
Table S2. Information of significant SNPs used for estimating the Linkage disequilibrium (LD) between PRKG1 and SCD. (XLSX 10 kb)
Additional file 3:
Table S3. 48 breeds for 1575 individuals in the database of 1000 Bull Genomes Project. (XLSX 9 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Shi, L., Lv, X., Liu, L. et al. A post-GWAS confirming effects of PRKG1 gene on milk fatty acids in a Chinese Holstein dairy population. BMC Genet 20, 53 (2019). https://doi.org/10.1186/s12863-019-0755-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12863-019-0755-7