Abstract
Background
People are paying more attention to the healthy and balanced diet with the improvement of their living standards. Milk fatty acids (FAs) have been reported that they were related to some atherosclerosis and coronary heart diseases in human. In our previous genome-wide association study (GWAS) on milk FAs in dairy cattle, 83 genome-wide significant single nucleotide polymorphisms (SNPs) were detected. Among them, two SNPs, ARS-BFGL-NGS-109493 and BTA-56389-no-rs associated with C18index (P = 0.0459), were located in the upstream of 1-acylglycerol-3-phosphate O-acyltransferase 3 (AGPAT3) gene. AGPAT3 is involved in glycerol-lipid, glycerol-phospholipid metabolism and phospholipase D signaling pathways. Hence, it was inferred as a candidate gene for milk FAs. The aim of this study was to further confirm the genetic effects of the AGPAT3 gene on milk FA traits in dairy cattle.
Results
Through re-sequencing the complete coding region, and 3000 bp of 5′ and 3′ regulatory regions of the AGPAT3 gene, a total of 17 SNPs were identified, including four in 5′ regulatory region, one in 5′ untranslated region (UTR), three in introns, one in 3′ UTR, and eight in 3′ regulatory region. By the linkage disequilibrium (LD) analysis with Haploview4.1 software, two haplotype blocks were observed that were formed by four and 12 identified SNPs, respectively. Using SAS9.2, we performed single locus-based and haplotype-based association analysis on 24 milk FAs in 1065 Chinese Holstein cows, and discovered that all the SNPs and the haplotype blocks were significantly associated with C6:0, C8:0 and C10:0 (P < 0.0001–0.0384). Further, with Genomatix, we predicted that four SNPs in 5′ regulatory region (g.146702957G > A, g.146704373A > G, g.146704618A > G and g.146704699G > A) changed the transcription factor binding sites (TFBSs) for transcription factors SMARCA3, REX1, VMYB, BRACH, NKX26, ZBED4, SP1, USF1, ARNT and FOXA1. Out of them, two SNPs were validated to impact transcriptional activity by performing luciferase assay that the alleles A of both SNPs, g.146704373A > G and g.146704618A > G, increased the transcriptional activities of AGPAT3 promoter compared with alleles G (P = 0.0004).
Conclusions
In conclusion, our findings first demonstrated the significant genetic associations of the AGPAT3 gene with milk FAs in dairy cattle, and two potential causal mutations were detected.
Similar content being viewed by others
Introduction
Milk fat is one of critical breeding objectives in dairy cattle. It is comprised of triglyceride (> 95%), diglyceride (2%), phospholipids (1%), cholesterol (0.05%) and small amount of free fatty acids (FAs) (~ 0.1%) [1]. The main components of triglyceride are glycerin and FAs, in which, the FAs act as precursors for the formation of other aroma components, such as esters and alcohols [2]. For the various milk fatty acid traits in Holstein cows, the estimated heritability values have been reported to be 0.14–0.33 for saturated fatty acids (SFAs) and 0.08–0.29 for unsaturated fatty acids (UFAs) [3,4,5,6,7].
Genome-wide association study (GWAS) is a commonly used strategy to identify potential genetic variants underlying important complex traits in human and domestic animals. So far, some candidate genes and QTL regions for milk production traits have been detected with GWA studies in dairy cattle, such as DLGAP1, AP2B1, SCD, BTA11 (1.59–3.37 Mb), and BTA3 (70.34–73.69 Mb) [8,9,10,11,12,13]. In our previous GWAS for milk FAs in Chinese Holstein cows, 83 genome-wide significant single nucleotide polymorphisms (SNPs) were detected in total [12], in which, two SNPs (ARS-BFGL-NGS-109493 and BTA-56389-no-rs) associated with C18index (P = 0.0459), were located in the upstream of 1-acylglycerol-3-phosphate O-acyltransferase 3 (AGPAT3) gene. In addition, we performed a joint GWAS for milk FAs in combined Chinese and Danish Holstein populations and found that a chromosome-wide significant QTL region of 146.29–146.31 Mb on BTA1 was associated with C18:0 [13]. The AGPAT3 gene was nearby this region with approximately 400 kb. 1-acylglycerol-sn-glycero 3-phosphate acyltransferase (AGPAT), encoded by the AGPAT3 gene, is one of the isoforms of AGPATs [14] and is involved in the glycerolipid (ko00561) and glycerophospholipid metabolisms (ko00564), and phospholipase D signaling pathway (ko04072). Mammalian AGPAT catalyzed the acylation of lysophosphatidic acid to form the phosphatidic acid that was the precursor of all glycerplipids [14]. Therefore, it was implied that the AGPAT3 gene was a promising candidate gene for milk FA traits in dairy cattle. The purpose of the present study was to further detect whether the AGPAT3 gene had significant genetic effects on milk FAs in a Chinese Holstein cow population.
Materials and methods
Animals and phenotypic data
In this study, a total of 1065 Chinese Holstein cows were used as descripted in a previous research [15], which milk samples were collected in Beijing Dairy Cattle Center (www.bdcc.com.cn) to measure milk FA contents. With the gas chromatography method, a total of 16 milk FAs (C6:0, C8:0, C10:0, C11:0, C12:0, C13:0, C14:0, C14:1, C15:0, C16:0, C16:1, C17:0, C17:1, C18:0, C18:1cis-9 and C20:0) were measured as the weight proportion of total fat weight [12]. With the phenotypes, we calculated five indices based on the formula\( \frac{cis\hbox{-} 9\mathrm{unsaturated}}{cis\hbox{-} 9\mathrm{unsaturated}+\mathrm{saturated}}\times 100 \) [16]: C14index = \( \frac{\mathrm{C}14:1}{\mathrm{C}14:1+\mathrm{C}14:0}\times 100 \), C16index = \( \frac{\mathrm{C}16:1}{\mathrm{C}16:1+\mathrm{C}16:0}\times 100 \), C17index = \( \frac{\mathrm{C}17:1}{\mathrm{C}17:1+\mathrm{C}17:0}\times 100 \), C18index = \( \frac{cis\hbox{-} 9}{\mathrm{C}18:1 cis\hbox{-} 9+\mathrm{C}18:0}\times 100 \), and total index = \( \frac{\mathrm{C}14:1+\mathrm{C}16:1+\mathrm{C}17:1+\mathrm{C}18:1 cis\hbox{-} 9}{\mathrm{C}14:1+\mathrm{C}14:0+\mathrm{C}16:1+\mathrm{C}16:0+\mathrm{C}17:1+\mathrm{C}17:0+\mathrm{C}18:1 cis\hbox{-} 9+\mathrm{C}18:0}\times 100 \). In addition, the summarized SFA and UFA, and SFA/UFA were obtained as well.
SNP identification and genotyping
Based on the genomic sequence of bovine AGPAT3 gene (Gene ID: 506607), 14 pairs of primers (Table S1) were designed by the Primer 3 version 4.0 (http://bioinfo.ut.ee/primer3-0.4.0/) and were synthesized in the Beijing Genomics Institute (Beijing, China) to amplify all the exons with partial adjacent intron region, and 3000 bp of 5′ and 3′ regulatory regions. As previously descripted [15], two DNA pools were constructed and the polymerase chain reaction (PCR) amplifications were performed with each DNA pool as template. To identify potential polymorphisms, the PCR amplification products were bi-directionally sequenced with an ABI3730XL DNA analyzer (Applied Biosystems, Foster, CA, USA). Then, the identified SNPs were genotyped for the 1065 cows by the matrix-assisted laser-desorption/ionization time of flight mass spectrometry (MALDI-TOF MS, Sequenom MassARRAY, Bioyong Technologies Inc., HK).
Linkage disequilibrium (LD) and association analyses
We estimated the LD among the identified SNPs of AGPAT3 gene with Haploview 4.1 (Broad Institute, Cambridge, MA, USA).
For association analysis, the 1065 cows were traced back to three-generation pedigrees to construct the kinship matrix (A-matrix) by SAS 9.2 (SAS institute, Cary, NC, USA), so that 3335 individuals were totally included. Single-locus and haplotype-based associations with 24 kind of milk FAs were performed by the following mixed animal model with SAS 9.2:
Here, Yijklm is the phenotypic value of each milk fatty acid trait; μ is the overall mean; Gi is the fixed effect corresponding to the genotype or haplotype combination of individual i; hj (j = 1–23) and lk (k = 1–4) were the fixed effect of farm j and stage of lactation ll, respectively; al is the random polygenic effect; Mm (m = 1–293) is the fixed effect of age at calving m; b is the regression coefficient of covariate M; and eijklm is the random residual. Further, we calculated the additive effect (a), dominant effect (d), and allele substitution effect (α) according to \( a=\frac{AA- BB}{2},d= AB-\frac{AA+ BB}{2}, and\alpha =a+d\left(q-p\right) \) [17]. Here, AA, AB and BB were the least square means of milk FAs corresponding to the genotypes, and p and q were the frequencies of allele A and B, respectively.
Prediction of changes of transcription factor binding sites (TFBSs) caused by the SNPs in 5′ regulatory region
We used the Genomatix software suite v3.9 (http://www.genomatix.de/cgi-bin/welcome/welcome.pl?s=d1b5c9a9015b02bb3b1a806f9c03293f) [18] to predict whether the four SNPs in 5′ regulatory region of AGPAT3 (g. 146702957G > A, g.146704373A > G, g.146704618A > G, and g.146704699G > A) changed the TFBSs.
Recombinant plasmid construction and luciferase assay
To detect the allele-specific effects of the SNPs g.146702957G > A, g.146704373A > G, g.146704618A > G, and g.146704699G > A on the transcriptional activity of AGPAT3 gene, eight luciferase reporter gene fragments (G and A of g. 146702957G > A; A and G of g.146704373A > G; A and G of g.146704618A > G; and G and A of g.146704699G > A) corresponding to the eight alleles of the four SNPs (Fig. 1a) were designed and synthesized (Genewiz, Suzhou, China). The eight fragments with the KpnI and Nhel restriction sites at the 5′ and 3′ termini, respectively, were cloned into the pGL4.14 luciferase assay vector (Promega, Madison, USA). In addition, all the plasmids were purified by the Endo-free Plasmid DNA Mini Kit II (OMEGA, omega bio-tek, Norcross, Georgia, USA), and were sequenced to confirm the integrity of each construct’s insertion.
Luciferase assay. a Sketches of recombinant plasmids with g.146702957G > A, g.146704373A > G, g.146704618A > G, and g.146704699G > A in the 5′ flanking region of AGPAT3gene. The nucleotides in red highlight refer to the SNP. b Luciferase assay analysis of the recombinant plasmids in HEK293 cells. Blank: Blank cells. PGL4.14: Empty vector. **: P < 0.01
The human embryonic kidney (HEK) 293 T cells were cultured with Dulbecco′s modified Eagle′s medium (Gibco, Life Technologies) and 10% fetal bovine serum (Gibco) at 37 °C in a humidified incubator containing 5% CO2. Before transfection, about 2 × 105 cells were seeded in each 24-well plate. For eight luciferase reporter gene fragments of g. 146702957G > A, g.146704373A > G, g.146704618A > G and g.146704699G > A, 500 ng constructed plasmid was co-transfected along with 10 ng pRL-TK Renilla luciferase reporter vector (Promega) into each well. All the experiments were performed in three replicates for each construct. Approximate 48 h after transfection, the cells were harvested and the activity of both firefly and Renilla luciferases were measured with a Dual-Luciferase Reporter Assay System (Promega) on a Modulus microplate multimode reader (Turner Biosystems, CA, USA). The average statistic of three replicates were calculated as the normalized luciferase data (Firefly/Renilla).
Results
Identification of SNPs
A total of 17 SNPs of the AGPAT3 gene was detected in this study (Table 1), which consisted of four (g.146702957G > A, g.146704373A > G, g.146704618A > G and g.146704699G > A) in 5′ flanking region, one (g.146705692G > A) in 5′ untranslated region (UTR), three (g.146725085 T > C, g.146726096A > G and g.146729107A > C) in introns, one (g.146735090G > T) in 3′ UTR, and eight (g.146737188C > T, g.146737545G > A, g.146737748 T > C, g.146737849C > T, g.146737879 T > G, g.146737916 T > C, g.146737946C > T and g.146738055G > A) in 3′ flanking region. The genotype and allele frequencies of the identified SNPs were presented in Table 1.
Estimation of LD among the identified SNPs of AGPAT3
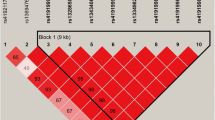
We used the haploview 4.1 to estimate the LD among these 17 SNPs, and observed two haplotype blocks (Fig. 2) that was formed by four and 12 SNPs, respectively. The haplotype block 1 included four haplotype combinations, namely, H1: GAAG (38%), H2: GAAA (32.2%), H3: AGGG (26.6%), and H4: GAGG (3%), and the haplotype block 2 had six haplotype combinations: H1 = GTAAGCGTCTTC, H2 = GCACGTACTGCT, H3 = GCAATCGTCTTC, H4 = ACACGCGTCTTC, H5 = GTGATCGTCTTC, and H6 = GCAAGCGTCTTC with their frequencies of 20%, 39.8%, 13.4%, 13.4%, 7.9% and 4.1%.
Linkage disequilibrium (LD) among the 17 SNPs of AGPAT3 gene. The blocks indicate haplotype blocks, and the text above the horizontal number is the SNP names. The values in boxes are pairwise SNP correlations (D´), while the bright red boxes without numbers inffer complete LD (D´ = 1). The boxes have the greater LD with the brigther red
Associations between AGPAT3 and milk FAs
The associations of the 17 SNPs with 24 milk FAs were summarized in Table 2. Among these SNPs, 17 were strongly associated with C6:0 (P < 0.0001–0.0004) and C8:0 (P < 0.0001–0.0384); 14 were significantly associated with total index (P < 0.0001–0.0318); ten were significantly associated with C10:0 (P = 0.0016–0.0151); nine were strongly associated with C17:1 (P < 0.0001–0.0149); seven were significantly associated with C20:0 (P < 0.0001–0.0072); five had significant associations with C14:0 (P < 0.0001–0.0477); five were strongly associated with C17index (P = 0.0006–0.0389); five had strong associations with C18:1cis-9 (P < 0.0001–0.0258); three had significant associations with C18:0 (P = 0.0020–0.0246); three had strong associations with SFA (P < 0.0001–0.0434); two were significantly associated with C17:0 (P = 0.0212–0.0413); two were significantly associated with UFA (P < 0.0001 and P = 0.0386); one was strongly associated with C18index (P = 0.0249); and one had significant association with SFA/UFA (P = 0.0005). However, no significant association was found with C11:0, C12:0, C13:0, C14:1, C15:0, C16:0, C16:1, C14index and C16index (P > 0.05).
Further, the additive (a), dominant (d), and allele substitution effects (α) of the 17 SNPs on each kind of fatty acid were calculated. Results showed that the 17 SNPs exhibited significant additive, dominant, and allele substitution effects on C6:0, C8:0, C10:0, C14:0, C16:0, C16:1, C17:0, C18:0, C18:1cis-9, C18index, C20:0, C14index, C16index, C17index, SFA, UFA, and total index (Table S2; P < 0.05). For C11:0, C12:0, C13:0, C14:1 and C15:0, none of significant additive, dominant, and allele substitution effects was found (P > 0.05).
Also, association analysis on two haplotype blocks with 24 milk FAs was performed (Table 3). The haplotype blcok1 was significantly associated with C6:0, C8:0, C10:0, C14:0, C18:0, C20:0, C17index and total index (P < 0.0001–0.0245), and the block 2 was strongly associated with C6:0, C8:0, C10:0, C14:0, C18:0, C17:1, C18:1cis-9, C18index, C20:0, C16index, C17index, SFA, UFA and total index (P < 0.0001–0.0498; Table 3). While, no significant association was found for C11:0, C12:0, C13:0, C14:1, C15:0, C16:0, C16:1, C14index and SFA/UFA (P > 0.05).
Prediction of TFBSs changing caused by the SNPs in 5′ regulatory region
By performing Genomatix software suite v3.9, it was predicted that that four SNPs in the 5′ regulatory region of AGPAT3 gene, g.146702957G > A, g.146704373A > G, g.146704618A > G and g.146704699G > A altered the binding sites of some transcription factors (Table 4). The allele A of g.146702957G > A created a TFBS for SMARCA3 (SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 3) and REX1 (REX1 transcription factor; zinc finger protein 42), respectively, and the allele G created a TFBS for VMYB (v-Myb, variant of AMV v-myb). The alleles A and G of g.146704373A > G created a TFBS for BRACH (Brachyury) and NKX26 (NK2 homeobox 6, Csx2), respectively. The allele G of g.146704618A > G created two TFBSs for ZBED4 (Zinc finger, BED-type containing 4; GC-box binding sites) and SP1 (Stimulating protein 1, ubiquitous zinc finger transcription factor). The allele G of g.146704699G > A created two TFBSs for USF1 (Upstream stimulating factor 1) and ARNT (AhR nuclear translocator homodimers), and the allele A created a TFBS for FOXA1 (Forkhead box protein A1, hepatocyte nuclear factor 3-alpha (HNF-3-alpha)).
Exploring for luciferase activity altered by the SNPs in 5′ regulatory region
To validate the TFBS prediction results, the luciferase assay was further performed for the four SNPs (g.146702957G > A, g.146704373A > G, g.146704618A > G and g.146704699G > A) (Fig. 1b). We observed that the luciferase activities of six constructs containing g.146704373A > G, g.146704618A > G, and g.146704699G > A, were significantly higher than that of the pGL4.14 empty vector (P < 0.0007) and blank control (P < 0.0008), while g.146702957G > A did not (P > 0.05). Further, the luciferase activity of alleles A of g.146704373A > G and g.146704618A > G were significantly higher than that of their alleles G (P = 0.0004; Fig. 1b). The luciferase activity of allele G of g.146704699G > A was higher than that of allele A, while not significant (P > 0.05). These results indicated that the transcriptional activity of the AGPAT3 gene significantly altered by g.146704373A > G and g.146704618A > G might be the reasons strongly impacted on FAs.
Discussion
This study was a follow-up investigation for our previous GWAS on milk FAs in Chinese Holstein [12]. AGPAT3 is involved in pathways related to lipid metabolism (ko00561, ko00564 and ko04072). In human, docosapentaenoic acid as the substrate of AGPAT3 protein transfers a fatty acid in sn-2 position of lysophosphatic acid, a step in the phospholipid biosynthesis pathway [19]. Here, we detected that the AGPAT3 gene mainly impacted the medium-chain milk FAs in dairy cattle.
Mammalian AGPAT catalyzed the acylation of lysophosphatidic acid to form the phosphatidic acid, which was the precursor of all glycerplipids [14]. For the AGPAT families, AGPAT1 had significant association with milk FA CLA [20], and AGPAT6 was strongly associated with C14:0, C16:0, C10:1, C12:1, C14:1 and C16:1 [21]. In our previous GWA studies [12, 13], AGPAT3 gene was identified as a candidate for two milk FAs, C18index and C18:0. In this study, using an independent Chinese Holstein population that was different from the previous GWA studies, we also observed that AGPAT3 showed a significant genetic effect on C18index and C18:0. In addition, our results revealed that the AGPAT3 had strong associations with C6:0, C8:0 and C10:0. Overall, the previous GWASs and this study suggested that AGPAT3 gene had significantly genetic effects on milk FAs.
Sequences-specific binding of transcription factors to the regulatory regions on the DNA is a key regulatory mechanism that affects gene expression and hence heritable phenotypic variation [22, 23]. Eukaryotic regulatory sequences, including enhancers and promoters, are typically between a hundred and several thousand base pairs in length, and can harbor many TFBSs [24]. It is essential to understand the evolution dynamics of transcription factor binding for understanding the evolution of gene regulation [25]. In this study, by prediction, g.146704373A > G changed the bindings of transcription factors (TFs) BRACH and NKX26, and g.146704618A > G altered the bindings of TFs ZBED4 and SP1. Further, we used the luciferase assay to verify that the alleles A of g.146704373A > G and g.146704618A > G strongly increased the transcription activity of the AGPAT3 gene than the alleles G. Previous studies showed that BRACH as a regulatory factor directly activated downstream mesoderm-specific genes to exert its mesoderm-inducing effects [26], and NKX26 restrained the transcription activity of Cx40 through the F151L missense mutation to impact the heart development [27]. ZBED4 could act as a co-repressor of nuclear hormone receptors (NHRs) by its LXXLL motifys in cones [28]. Through interfering with the recruitment of SP1 to ZNF132 promoter region, methylation of SP1-binding site can inhibit ZNF132 transcriptional expression to impact the tumor in the development of esophageal squamous cell carcinoma [29]. These reports have indicated that the TFs BRACH, NKX26, ZBED4 and SP1 could activate or repress the expression of their target genes. Based on our association analysis, the cows with the AA genotypes of g.146704373A > G and g.146704618A > G of AGPAT3, yielded significantly lower contents of C6:0 and C8:0 than those with GG genotypes. According to above, we deduced that the BRACH as a TF might activate AGPAT3 gene transcription activity by binding to the allele A of g.146704373A > G thereby reducing the contents of C6:0 and C8:0, while, the transcription factors NKX26, ZBED4 and SP1 might have the contrary effects.
Nowadays, genomic selection is the main implication for dairy cattle breeding, where the genomic chips are used. Among the SNP markers in these chips, most of them were collected from the current SNP database and almost evenly distributed across the whole genome. Hence, g.146704373A > G and g.146704618A > G of AGPAT3 as the potentially causal mutations could be put into the SNP chip instead of used in marker selection to increase selection efficiency in some specific dairy cattle populations to improve the contents of milk FAs.
Conclusion
In conclusion, through a post-GWAS approach, our study firstly indicated there were significant genetic associations between the AGPAT3 gene and milk FAs in dairy cattle. Further, we found that two SNPs in 5′ regulatory region (g.146704373A > G and g.146704618A > G) changed the transcriptional activity of AGPAT3 implying their potential causal function. These findings provided important molecular information for dairy cattle breeding.
Availability of data and materials
All relevant data are available within the article and its supplementary information.
Abbreviations
- a:
-
Additive
- AGPAT:
-
1-Acylglycerol-sn-glycero 3-phosphate acyltransferase
- AGPAT3:
-
1-Acylglycerol-3-phosphate O-acyltransferase 3
- ARNT:
-
AhR nuclear translocator homodimers
- BRACH:
-
Brachyury
- d:
-
Dominant
- FA:
-
Fatty acid
- FOXA1:
-
Forkhead box protein A1, hepatocyte nuclear factor 3-alpha (HNF-3-alpha)
- GWAS:
-
Genome-wide association study
- HEK:
-
Human embryonic kidney
- LD:
-
Linkage disequilibrium
- NKX26:
-
NK2 homeobox 6, Csx2
- PCR:
-
Polymerase chain reaction
- REX1:
-
REX1 transcription factor; zinc finger protein 42
- SFA:
-
Saturated fatty acids
- SMARCA3:
-
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 3
- SNP:
-
Single nucleotide polymorphism
- SP1:
-
Stimulating protein 1, ubiquitous zinc finger transcription factor
- TFBS:
-
Transcription factor binding site
- UFA:
-
Unsaturated fatty acid
- USF1:
-
Upstream stimulating factor 1
- UTR:
-
Untranslated region
- VMYB:
-
v-Myb, variant of AMV v-myb
- ZBED4:
-
Zinc finger, BED-type containing 4, GC-box binding sites
- α:
-
Substitution
References
Staniewski B, Kielczewska K, Smoczynski M, Baranowska M, Czerniewicz M, Brandt W. Effect of high pressures on the composition of milk fat triacylglycerols. Milchwissenschaft. 2012;67(1):18–21.
Delafuente MA, Fontecha J, Juarez M. Fatty-acid composition of the triglyceride and free fatty-acid fractions in different cows-milk, ewes-milk and goats-milk cheeses. Z Lebensm Unters For. 1993;196(2):155–8.
Krag K, Poulsen NA, Larsen MK, Larsen LB, Janss LL, Buitenhuis B. Genetic parameters for milk fatty acids in Danish Holstein cattle based on SNP markers using a Bayesian approach. BMC Genet. 2013;14:79.
Petrini J, Iung LH, Rodriguez MA, Salvian M, Pertille F, Rovadoscki GA, et al. Genetic parameters for milk fatty acids, milk yield and quality traits of a Holstein cattle population reared under tropical conditions. J Anim Breed Genet. 2016;133(5):384–95.
Narayana SG, Schenkel FS, Fleming A, Koeck A, Malchiodi F, Jamrozik J, et al. Genetic analysis of groups of mid-infrared predicted fatty acids in milk. J Dairy Sci. 2017;100(6):4731–44.
Stoop WM, Schennink A, Visker MH, Mullaart E, van Arendonk JA, Bovenhuis H. Genome-wide scan for bovine milk-fat composition. I. Quantitative trait loci for short- and medium-chain fatty acids. J Dairy Sci. 2009;92(9):4664–75.
Schennink A, Stoop WM, Visker MH, van der Poel JJ, Bovenhuis H, van Arendonk JA. Short communication: Genome-wide scan for bovine milk-fat composition. II. Quantitative trait loci for long-chain fatty acids. J Dairy Sci. 2009;92(9):4676–82.
Chen Z, Yao Y, Ma P, Wang Q, Pan Y. Haplotype-based genome-wide association study identifies loci and candidate genes for milk yield in Holsteins. PLoS One. 2018;13(2):e0192695.
Palombo V, Milanesi M, Sgorlon S, Capomaccio S, Mele M, Nicolazzi E, et al. Genome-wide association study of milk fatty acid composition in Italian Simmental and Italian Holstein cows using single nucleotide polymorphism arrays. J Dairy Sci. 2018.
Knutsen TM, Olsen HG, Tafintseva V, Svendsen M, Kohler A, Kent MP, et al. Unravelling genetic variation underlying de novo-synthesis of bovine milk fatty acids. Sci Rep. 2018;8(1):2179.
Yue SJ, Zhao YQ, Gu XR, Yin B, Jiang YL, Wang ZH, et al. A genome-wide association study suggests new candidate genes for milk production traits in Chinese Holstein cattle. Anim Genet. 2017;48(6):677–81.
Li C, Sun D, Zhang S, Wang S, Wu X, Zhang Q, et al. Genome wide association study identifies 20. novel promising genes associated with milk fatty acid traits in Chinese Holstein. Plos One. 2014;9(5):e96186.
Li X, Buitenhuis AJ, Lund MS, Li C, Sun D, Zhang Q, et al. Joint genome-wide association study for milk fatty acid traits in Chinese and Danish Holstein populations. J Dairy Sci. 2015;98(11):8152–63.
Lu B, Jiang YJ, Zhou Y, Xu FY, Hatch GM, Choy PC. Cloning and characterization of murine 1-acyl-sn-glycerol 3-phosphate acyltransferases and their regulation by PPARalpha in murine heart. Biochem J. 2005;385(Pt 2):469–77.
Shi L, Liu L, Ma Z, Lv X, Li C, Xu L, et al. Identification of genetic associations of ECHS1 gene with milk fatty acid traits in dairy cattle. Anim Genet. 2019;50(5):430–8.
Kelsey JA, Corl BA, Collier RJ, Bauman DE. The effect of breed, parity, and stage of lactation on conjugated linoleic acid (CLA) in milk fat from dairy cows. J Dairy Sci. 2003;86(8):2588–97.
Falconer DS, Mackay TFC. Introduction to quantitative genetics. 1996: Ed. 4 xv + 464 pp.
Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21(13):2933–42.
Lemaitre RN, Tanaka T, Tang W, Manichaikul A, Foy M, Kabagambe EK, et al. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE consortium. PLoS Genet. 2011;7(7):e1002193.
Gebreyesus G, Buitenhuis AJ, Poulsen NA, Visker M, Zhang Q, van Valenberg HJF, et al. Multi-population GWAS and enrichment analyses reveal novel genomic regions and promising candidate genes underlying bovine milk fatty acid composition. BMC Genomics. 2019;20(1):178.
Bouwman AC, Bovenhuis H, Visker MH, van Arendonk JA. Genome-wide association of milk fatty acids in Dutch dairy cattle. BMC Genet. 2011;12:43.
Tugrul M, Paixao T, Barton NH, Tkacik G. Dynamics of transcription factor binding site evolution. PLoS Genet. 2015;11(11):e1005639.
Talebzadeh M, Zare-Mirakabad F. Transcription factor binding sites prediction based on modified. nucleosomes. Plos One. 2014;9(2):e89226.
Yao P, Lin P, Gokoolparsadh A, Assareh A, Thang MW, Voineagu I. Coexpression networks identify brain region-specific enhancer RNAs in the human brain. Nat Neurosci. 2015;18(8):1168–74.
Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328(5981):1036–40.
Chen M, Wu Y, Zhang H, Li S, Zhou J, Shen J. The roles of embryonic transcription factor BRACHYURY in tumorigenesis and progression. Front Oncol. 2020;10:961.
Heathcote K, Braybrook C, Abushaban L, Guy M, Khetyar ME, Patton MA, et al. Common arterial trunk associated with a homeodomain mutation of NKX2.6. Hum Mol Genet. 2005;14(5):585–93.
Farber DB, Theendakara VP, Akhmedov NB, Saghizadeh M. ZBED4, a novel retinal protein expressed in cones and Müller cells. Adv Exp Med Biol. 2010;664:79–87.
Jiang D, He ZL, Wang CJ, Zhou YH, Li F, Pu WL, et al. Epigenetic silencing of ZNF132 mediated by methylation-sensitive Sp1 binding promotes cancer progression in esophageal squamous cell carcinoma. Cell Death Dis. 2018;10(1):1.
Acknowledgements
We appreciate Beijing Dairy Cattle Center and Beijing Sanyuanlvhe Dairy Farming Center for providing the milk, blood, and semen samples of Chinese Holstein.
Funding
This work was financially supported by the National Natural Science Foundation of China (31872330, 31802041), Beijing Science and Technology Program (20200105, D171100002417001), earmarked fund for Modern Agro-industry Technology Research System (CARS-36), and the Program for Changjiang Scholar and Innovation Research Team in University (IRT_15R62).
Author information
Authors and Affiliations
Contributions
DS and YY conceived and designed the experiments, LL prepared the DNA samples for SNP identification and genotyping with the help of XW, ZM, XL, YL, and FZ, XL measured the phenotypes of milk fatty acids, LS and XW analyzed the data, and LS, BH and DS prepared the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All protocols for collection of the samples of experimental individuals and phenotypic observations were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at China Agricultural University (Permit Number: DK996). Milk, blood and semen samples were collected specifically for this study following standard procedures with the full agreement of the Beijing Sanyuanlvhe Dairy Farming Center who owned the Holstein cows and bulls, respectively.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Supplementary Information
Additional file 1: Table S1.
PCR primers information of AGPAT3 gene
Additional file 2: Table S2.
Additive (a), dominant (d) and allele substitution (α) effects of 17 SNPs on milk fatty acid traits of AGPAT3 gene in Chinese Holstein cows
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Shi, L., Wu, X., Yang, Y. et al. A post-GWAS confirming the genetic effects and functional polymorphisms of AGPAT3 gene on milk fatty acids in dairy cattle. J Animal Sci Biotechnol 12, 24 (2021). https://doi.org/10.1186/s40104-020-00540-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40104-020-00540-4