Abstract
Background
The initiation of flowering transition in rice (Oryza sativa) is a complex process regulated by genes and environment. In particular, drought can interfere with flowering; therefore, many plants hasten this process to shorten their life cycle under water scarcity, and this is known as drought-escape response. However, rice has other strategies; for example, drought stress can delay flowering instead of accelerating it. RICE CENTRORADIALIS 1 (RCN1) is a TERMINAL FLOWER-like gene that influences rice flowering transition and spike differentiation. It interacts with 14–3-3 proteins and transcription factor OsFD1 to form a florigen repression complex that suppresses flowering transition in rice.
Results
In this study, we explored the role of RCN1 in the molecular pathway of drought-regulated flowering transition. The rcn1 mutant plants displayed early heading under both normal water and drought stress conditions, and they were more insensitive to drought stress than the wild-type plants. Abscisic acid (ABA) signaling-mediated drought-induced RCN1 is involved in this process.
Conclusions
Thus, RCN1 plays an important role in the process of drought stress inhibiting flowering transition. It may worked by suppressing the protein function rather than transcription of HEADING DATE 3a.
Similar content being viewed by others
Background
Drought is one of the most severe natural stresses affecting crops. Owing to inadequate irrigation plans, at least 30% of the total rice cultivation area worldwide is rainfed (Dixit et al. 2014). Because water resources are unevenly distributed in terms of both space and time, rice fields are severely affected by uneven rainfall, which leads to seasonal water shortages (Wu et al. 2011). Rice, which is consumed by more than half of the global population, has been grown under flooded irrigation conditions over the course of its evolution as a food crop because it requires large quantities of water and is sensitive to water-deficient soils. Approximately 3000–5000 L of water is used to produce 1 kg of rice, and this is almost twice the amount needed to grow other crops such as maize and wheat (Dixit et al. 2014). Drought can lead to a considerable delay in rice plant growth and spikelet development, thereby resulting in low crop yield, which causes substantial economic losses. Thus, understanding the molecular mechanisms underlying the effects of drought on key developmental processes in rice is crucial for the production and breeding selection of this crop.
Floral transition marks the switch from vegetative growth to reproductive growth. The timing of floral transition is a key determinant of crop production and adaptability to complex environmental conditions. The effects of drought on flowering time have been documented in several plant species (Bernal et al. 2011; Bocco et al. 2012; Franks 2011; Riboni et al. 2013; Riboni et al. 2016; Sherrard and Maherali 2006). One strategy of plants to endure water scarcity is to accelerate flowering transition, and this is known as drought-escape (DE) response. This response enables plants to accelerate the completion of their life cycle and produce seeds before the stress conditions become too severe (Franks 2011; Riboni et al. 2013; Riboni et al. 2016). The molecular mechanisms underlying the regulation of flowering time under drought stress have recently been elucidated in Arabidopsis. In this species, the positive regulator GIGANTEA (GI) and the florigen genes FLOWERING LOCUS T (FT) and TWIN SISTER OF FT (TSF) play key roles in regulating the DE response, which occurs only under long-day (LD) growth conditions. Abscisic acid (ABA)-deficient mutants, aba1 and aba2, flowered later than wild type (WT) plants and showed a reduced DE response under LD conditions (Riboni et al. 2016), implying that drought stress positively regulates the transcription of these florigens in an ABA-dependent manner (Riboni et al. 2013; Riboni et al. 2016).
In addition, ABA has been found to repress flowering independent of florigen genes. The flowering-promoting gene SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) was repressed by ABA in the shoot apical meristem (Riboni et al. 2016). Rice plants delay flowering under drought stress to avoid reproductive growth under adverse conditions (Bocco et al. 2012; Fukai et al. 1999; Galbiati et al. 2016; Zhang et al. 2006). Drought stress strongly reduces the transcription of EARLY HEADING DATE 1 (Ehd1), which is upstream of florigen genes, and accumulation of HEADING DATE 3a (Hd3a) and RICE FLOWERING LOCUS T1 (RFT1), and ultimately leads to delayed flowering (Galbiati et al. 2016). However, a recent study reported that rice responds to drought differently, depending on its intensity, suggesting that a less severe drought condition could trigger the DE response (Du et al. 2018). The drought response of rice depends on the ABA-signaling pathway, similar to that of Arabidopsis. Furthermore, the regulation of rice flowering under drought conditions is not dependent on the photoperiod, unlike that of Arabidopsis (Zhang et al. 2016). Early heading date 1 (Ehd1) is specific for flowering in rice (Doi et al. 2004), and it plays a key role in integration of drought stress and the photoperiodic signals (Galbiati et al. 2016). Grain number, plant height, and heading date 7 (Ghd7) is another specific gene in rice, which involved in the regulation of DE (Du et al. 2018; Wu et al. 2008). OsGIGANTEA (OsGI) does not integrate photoperiodic and drought signals as in Arabidopsis (Galbiati et al. 2016). Indicating that the molecular mechanism underlying flowering initiation differs between these two species.
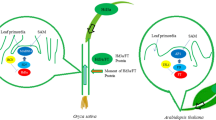
Florigen is the ultimate regulator of flowering under varied environmental conditions. It belongs to the family of phosphatidylethanolamine-binding proteins (PEBPs). TERMINAL FLOWER 1 (TFL1), an anti-florigen, which also belongs to this family, suppresses flowering and thus opposes the actions of FT in mediating flowering transition, although they are expressed in a similar pattern and have similar structures (Guo et al. 2014; Hanano and Goto 2011). RICE CENTRORADIALIS 1 (RCN1), a rice TFL1 homolog, performs functions similar to those of TFL1 in the flowering regulation process (Nakagawa et al. 2002; Zhang et al. 2005). A genome-wide association study of a population of 950 rice varieties collected worldwide identified RCN1 as an important locus associated with flowering time (Huang et al. 2012). The underlying molecular mechanism of RCN1 has been studied recently. The RCN1 protein is synthesized in the leaves and transported to the shoot apical meristem (SAM), where it opposes the action of florigen in floral transitions by competing with it to form complexes with the 14–3-3 and OsFD1 proteins (Kaneko-Suzuki et al. 2018). In addition, the genome-wide association study found that RCN1, located within one quantitative trait locus (q12), is related to drought tolerance during the vegetative phase (Hoang et al. 2019). Nevertheless, the roles of RCN1 in the integration of environmental signals in rice are poorly understood.
In the present study, the mutation of RCN1 significantly advanced the heading date of rice. Moreover, we suggest an ABA-dependent response pathway for drought stress-induced RCN1 expression. In this pathway, two transcription factors, ABA-RESPONSIVE ELEMENT BINDING PROTEIN 1 (OsAREB1) and ABA RESPONSIVE ELEMENT (ABRE)-BINDING FACTOR (OSBZ8), are induced within minutes upon exposure to ABA and drought stresses and both function as trans-activators that modulate the drought response of RCN1.
Results
Influence of Drought on the Heading Date Is Weaker in rcn1 Mutants than in WT Plants
To test if RCN1 loss of function affects rice flowering time, we obtained two different insert mutations in the coding region of RCN1 using the clustered regularly interspaced short palindromic repeats-associated protein-9 (CRISPR-Cas9) technology (Figure S1), the frame-shift mutations caused premature termination of translation or amino acid substitutions. In both mutant lines RCN1 was not functional. The experiment was carried out during the growing season. Rice plants were subject to severe drought stress, as the soil relative water capacity (SRWC) was maintained between 30%–50% (Figure S2). In rcn1 mutants, heading occurred approximately 7 days earlier than that in the WT plants under normal water conditions (Table 1), indicating a negative role of RCN1 in controlling flowering transition. Both WT and rcn1 mutant plants showed a significantly late-heading phenotype under severe drought conditions (Table 1). It is noteworthy that the heading date of the WT plants was delayed for approximately 14 days, and the two rcn1 mutant plants showed delays of 9.2 and 10.9 days, respectively. The results indicated that RCN1 loss of function reduced the effect of drought on rice heading date (Table 1).
The expression of RCN1 in the rcn1 mutant plants was not significantly different from that of the wild-type plants under normal watering conditions (Figure S3a). In contrast, in rcn1 mutant plants, RCN1 showed a significantly decreased responsiveness to drought. The rice florigens Hd3a and RFT1 promote flowering. Under normal watering conditions, there was no obvious difference in Hd3a/RFT1 expression levels between rcn1 mutants and wild-type plants. Under drought stress conditions, both the wild-type and the rcn1 mutant plants showed a significant down-regulation of Hd3a and RFT1. However, Hd3a and RFT1 were not regulated by RCN1; they were down-regulated under drought conditions in an RCN1-independent manner (Figure S3b, c). Drought stress can delay flowering by inhibiting florigen expression, and there was no effect of RCN1 on florigens transcription.
Tissue-Specific Expression and Localization of RCN1
To examine the spatio–temporal expression of RCN1, a quantitative reverse transcription- PCR (qRT-PCR) analysis was performed. The expression of RCN1 was detected in the leaves, roots, and stems throughout the growth period, and the highest expression was observed in the roots (Fig. 1a). To further examine the expression pattern of RCN1, the 5′ distal region (4 kb) of the RCN1 locus was fused with the beta-glucuronidase (GUS) reporter and used to conduct histochemical staining of the GUS gene. The expression of GUS was detected in the vascular tissues of the leaves and roots (Fig. 1b, c). In the cross-section of stems, a high GUS expression was detected in the nodes and basal internodes (Fig. 1d, e), whereas no expression was detected in young panicles (Fig. 1f). These results were consistent with the qRT-PCR results. Furthermore, GUS expression was observed in the glumes, and this has never been reported (Fig. 1g).
Expression patterns of RCN1 in rice. Expression of RCN1 RNA in rice (a). RCN1 expression was analyzed by quantitative real-time PCR throughout the growth period. Actin RNA was used as the control. DAS: days after seeding. GUS staining images of pRCN1::GUS:NOS in transgenic rice leaf (b), root (c), stem node (d), basal internodes (e), young panicle (f), and glume (g). The localization of the RCN1-GFP protein was visualized in confocal images of the leaf (h) and root (i) in pRCN1::RCN1-GFP:NOS transgenic rice plants, and the subcellular localization of the RCN1-GFP protein was visualized lateral root cells (j). Bars = 100 μm (for b-c), 1 mm (for d-g)
To examine the localization of the RCN1 protein in the plant, we constructed an expression vector with GREEN FLUORESCENT PROTEIN (GFP) as the reporter gene, by fusing the RCN1-coding region with the GFP-coding region (RCN1-GFP), and used the same promoter as that in the GUS analysis. The distribution of RCN1-GFP was consistent with the results of the qRT-PCR and GUS staining, suggesting the localization of RCN1 to the vasculature of the leaves and roots (Fig. 1h, i). We also observed the subcellular localization of RCN1-GFP in lateral root cells, where it was mainly distributed in the cell nucleus, cytoplasm, and cellular membrane (Fig. 1j); the same was observed in rice protoplasts (Figure S4).
Drought Stress Activates RCN1 Transcription in an ABA-Dependent Manner
For the osmotic stress treatment, plants were transferred to pots containing three different concentrations of polyethylene glycol (PEG6000), while those grown without PEG6000 were used as the controls. The expression of RCN1 was detected by qRT-PCR. The results indicated that 30% PEG6000 strongly increased the expression of RCN1 in the shoots and roots (Fig. 2a, b), and therefore this concentration was used in the subsequent experiments. It is widely known that ABA is accumulated in response to drought; thus, experiments were designed to verify whether RCN1 would respond to ABA. We added 50 μM ABA to the rice culture solution and detected the expression level of RCN1 at 0, 3, 6, 24, 30, 48, 54, 96, and 102 h of incubation, by qRT-PCR. Overall, ABA strongly increased the expression of RCN1 in the shoots and roots, particularly in the latter (Fig. 2c, d). Because the upregulation of RCN1 in the shoots was only detected 24 h post-ABA treatment, samples collected at this time point were used to evaluate the effects of ABA treatment. To evaluate the relationship between ABA and RCN1, we further examined the effect of different concentrations of exogenous ABA on RCN1 expression. The results showed that RCN1 expression was proportional to the exogenous ABA concentration applied (Fig. 2e, f). Compared with the control, the expression of RCN1 in the shoots increased by up to 10-fold (Fig. 2e), far below the 39-fold observed in the roots (Fig. 2f). To identify if PEG6000 induced RCN1 expression in an ABA-dependent manner, we treated rice plants with 40 μΜ fluridone for 30 min, and then moved them to 30% PEG6000 solution with fluridone. Fluridone, an inhibitor of carotenoids, which are the main precursors of ABA in plants (Raikhel et al. 1986; Kowalczyk-Schroder and Sandmann 1992), also inhibits the biosynthesis of ABA. Fluridone treatment was effective inhibit the increase in ABA content (Hsu et al. 2006; HSU et al. 2003; Perales et al. 2005; Shi et al. 2012). The results revealed that, after blocking ABA synthesis, RCN1 did not respond to PEG6000 (Fig. 2g, h), suggesting that the PEG6000-mediated induction of RCN1 was dependent on ABA. The ABA-mediated induction of RCN1 was completely inhibited by cycloheximide (CHX), which is an inhibitor of protein synthesis (Fig. 2i, j). This indicated that protein synthesis is necessary to ABA regulate RCN1 expression, one reason for the requirement may have been that transcription factors play an important role in regulation. PEG6000- and ABA-induced RCN1 expression was partially restrained by K252a, which is a phosphorylation blocker (Fig. 2g, h, k, i). These results revealed an ABA-mediated induction of RCN1 that was dependent on phosphorylation, and provided further evidence for ABA-mediated induction of RCN1 under drought.
Expression levels of RCN1 in the leaves and roots under different PEG6000 treatment concentrations (a and b), different ABA treatment times (c and d), and ABA treatment concentrations (e and f). PEG6000-induced RCN1 levels in the leaves (g) and roots (h) depended on ABA synthesis and protein phosphorylation. ABA-induced RCN1 levels in the leaf (i) and root (j) depended on de novo protein synthesis. CHX: cycloheximide. ABA-induced RCN1 expression levels in the leaf (k) and (l) root depended on protein phosphorylation. Note: Statistical analysis was performed using the Student’s t test; significantly different values at P < 0.05 (*) or P < 0.01 (**) are indicated
ABA Induces RCN1 through Transcription Factors
The basic leucine-zipper (bZIP) transcription factors are bound to the ABA-responsive cis-element ABRE in the promoters of ABA-inducible genes to regulate plant stress responses. The conserved sequence of ABRE is ACGT, and the C/G/A nucleotides flanking the ACGT core enhance bZIP protein binding specificity and affinity (Hattori et al. 2002). We screened 26 ABA response transcription factors from authoritative databases and selected five transcription factors that accumulated in response to ABA faster than RCN1 (Figure S5a). A new ABA treatment experiment was then carried out to determine the expression levels of these transcription factors. Finally, OsAREB1, OSBZ8, and TRAB1 showed a rapid and strong response to ABA in both leaves and roots (Figure S5b, c). Because the induction of OsAREB1, OSBZ8, and TRAB1 expression by ABA was not blocked by CHX (Figure S5d, e), they appear to be primary ABA response genes.
Electrophoretic mobility shift assays (EMSA) were used to examine the binding activities of the OsAREB1, OSBZ8, and TRAB1 proteins with the ABRE element in the RCN1 promoter. The 14 sequence specific fragments around the RCN1 locus contained 19 ACGT elements that were synthesized and labeled with biotin (Fig. 3a). OsAREB1, OSBZ8, and TRAB1 were expressed and extracted from the BL21 competent E. coli strain. Among the six different fragments tested, OsAREB1 and OSBZ8 exhibited the strongest interaction with Probe 13 (containing three ACGT elements), a slightly weaker interaction with Probes 9, 10, and 12 (each containing two ACGT elements), and the weakest interactions with Probes 11 and 14 (each containing one ACGT element) (Fig. 3b, c). To examine the specificity of these interactions, a 50- or 200-fold unlabeled probe was applied to compete with the labeled probe. The interactions of OsAREB1 and OSBZ8 and labeled probes were inhibited by excess unlabeled probes. No interaction was detected with mutant probes that lacked the ABRE element sequence (Fig. 3b, c). There were no interactions between OsAREB1 and OSBZ8 and Probes 1–8 (Figure S6a, b), and between the TRAB1 protein and any of the 14 probes (Figure S6c). Therefore, OsAREB1 and OSBZ8 seemed to bind only to the ABRE element, which is located in the 3′ distal region of the RCN1 locus, and their binding ability is likely be related to the number of ABRE elements included in the DNA fragments.
The EMSA experiments revealed that OsAREB1 and OSBZ8 can bind to the 3′ distal region of the RCN1 locus in vitro. To determine whether they can regulate RCN1 expression in vivo, glucocorticoid-inducible transgenic lines carrying plasmid Gos2::OsAREB1 or Gos2::OSBZ8 in Nipponbare background were generated. The expression of OsAREB1 and OSBZ8 in the Gos2::OsAREB1 and Gos2::OSBZ8 transgenic plant leaves, respectively, was induced by a low concentration of dexamethasone (DEX) (Fig. 4a, b). As expected, the expression of RCN1 was upregulated with the increase in OsAREB1 and OSBZ8 expression (Fig. 4c, d). Taken together, these findings suggest that OsAREB1 and OSBZ8 function as transcriptional regulators that modulate the drought response of RCN1 via an ABA-dependent pathway.
To further examine whether the 5′ proximal region of the RCN1 locus response to ABA, we treated pRCN1::RCN1-GFP:NOS and pRCN1::GUS:NOS transgenic plants with ABA, and detected the expression levels of GUS and GFP and the endogenous expression level of RCN1 (Fig. 5). The expression of GFP and GUS was not induced by ABA, whereas the endogenous expression of RCN1 was significantly induced (Fig. 4b, c). The results of the qRT-PCR indicated that the ABA response element was not located on the 5′ proximal region of RCN1, but may be the ABRE elements on 3′ distal region of RCN1.
Differences in promoters and terminators in pRCN1::GUS:NOS and pRCN1::RCN1-GFP:NOS transgenic plants and wild-type (WT) plants (a). The expression levels of native RCN1 and GUS in the pRCN1::GUS:NOS transgenic plants under ABA treatment (b). The expression levels of native RCN1 and GFP in the pRCN1::RCN1-GFP:NOS transgenic plants under ABA treatment (c)
Discussion
RCN1 Mediates Drought-Delayed Heading
There are two possible drought stress response modes in rice: low drought stress in the early growth stage of rice triggers the DE response (Du et al. 2018) and severe drought stress delays flowering (Bocco et al. 2012; Fukai et al. 1999; Galbiati et al. 2016; Zhang et al. 2006). In Arabidopsis, drought stress promoted early flowering only under LD conditions (Riboni et al. 2013; Riboni et al. 2016). However, in rice, the heading time is affected by drought stress under both SD and LD conditions (Galbiati et al. 2016), although rice is a short-day plant. In the present study, when rice was subject to drought stress, the heading of WT plants was postponed compared with that of the normally watered controls. These data agree with previous experimental results, that is, the flowering-delayed phenotype was largely observed under drought stress. In addition, the rcn1 mutants displayed apparently earlier heading than the WT plants, implying a negative function for RCN1 in floral transition. The heading delay of the rcn1 mutants induced by drought was weaker than that of WT plants, suggesting that the effect of drought on the heading date of the rcn1 mutants was less pronounced. Previous studies have identified RCN1 as a negative floral gene, restraining floral transition by competing with Hd3a/RFT1 for 14–3-3 binding (Kaneko-Suzuki et al. 2018; Nakagawa et al. 2002; Zhang et al. 2005). A significant upregulation of RCN1 expression was observed under drought stress; thus, drought may inhibit Hd3a protein function by promoting the expression of RCN1. Previous studies suggested that the delayed flowering phenotype under drought stress was largely dependent on the downregulation of Hd3a (Galbiati et al. 2016). Both transcriptional regulation and protein function regulation contribute to the floral transition regulatory mechanism of drought.
Drought Induces RCN1 in an ABA-Dependent Manner
In the present study, we showed that drought regulates RCN1 expression through an ABA-dependent pathway. ABA is regarded as a signal that can transmit drought information when plants suffer drought stress. Under the ABA treatment, RCN1 expression showed faster and longer-lasting upregulation in the roots than in the leaves. The root is the first organ to perceive drought stress in the plant growth process, and ABA is then rapidly synthesized and transported upward to the leaf to transmit drought information. The sensitivity of RCN1 in the root to drought stress implies that it may play important roles in rice drought response and growth regulation. One of the ABA-mediated responses is the induced expression of a large number of genes, which are mediated by cis-regulatory elements, the ABREs. The presence of ABRE is essential for abiotic stress inducibility of an ABA-dependent pathway, and the transcription factor that binds to ABRE plays an important role in the expression of most ABA-response genes (Nakashima and Yamaguchi-Shinozaki 2013). Both OsAREB1 and OSBZ8 act as transcriptional regulators that modulate the ABA response of RCN1 by directly binding with the ABRE located in the 3′ distal region of the RCN1 locus. Transgenic Arabidopsis over-expressing OsAREB1 had higher resistance to drought and later flowering time (Jin et al. 2010). Thus, OsAREB1 could have the same function in rice. Our experiment showed the drought increased OsAREB1 and OSBZ8 expression in timing of flowering (Figure S7), thereby activating RCN1 expression, and then causing a flowering transition delay. Our study revealed that drought delayed the heading time partially by inducing RCN1 via an ABA signaling pathway, and that OsAREB1 and OSBZ8 are involved in this process.
RCN1 showed a decreased responsiveness to drought in rcn1 plants. Previous studies found that the Hd3a and RFT1 proteins can form a transcriptional repression complexes with BINDING REPRESSOR FACTOR 1 (HBF1) and HBF2 and then feedback to affect their own transcription in the leaves (Brambilla et al. 2017). As their homeotic gene, RCN1 might could also combine with a transcription factor to form a transcriptional complex to regulate itself in leaves.
RCN1 Protein Is Localized to the Nuclei of Leaf and Root Cells
In Arabidopsis, the long-distance transport modes of FT from the phloem to shoot apex have been identified (Corbesier et al. 2007; Liu et al. 2012). The FT-homologous genes Hd3a and RFT1 were strongly expressed in the leaves; these proteins moved from the leaf to the SAM, thereby inducing flowering (Tamaki et al. 2007; Taoka et al. 2011). As the antagonistic gene of florigen, RCN1 was expressed in the root, leaf, and stem of rice. In the rice leaf, RCN1 showed similar expression patterns to florigen, in the vascular bundles. The RCN1 protein was transported from the vascular tissue to the SAM (Kaneko-Suzuki et al. 2018). In the present study, RCN1 presented the highest expression in rice roots from the vegetative to late reproductive phases. The expression of RCN1 was relatively lower in the leaves and stems than in the roots. These results implied that RCN1 might have important functions in the root. In the present study, RCN1 was undetectable in the young panicle and the RCN1 promoter activity in gRCN-GUS plants was also undetected in the young panicle. This might be because RCN1 is expressed in the vascular bundle, which is not differentiated in the young panicle. Kaneko-Suzuki (2018) demonstrated the same result. We observed that the RCN1-GFP fusion protein was localized to the cytomembrane, cytoplasm, and nucleus. A previous study reported the localization of RFT1 in whole cells (Song et al. 2017) and Hd3a in the cytoplasm and nucleus (Taoka et al. 2011). The nuclear localization of RCN1, Hd3a, and RFT1 was related to their transcription repression/activation functions that were achieved in combination with the transcription factor. Our results suggest that RCN1 is synthesized outside the SAM in the flowering transition period to regulate flowering transition and branch differentiation. As a movement protein, RCN1 may have a signal transmission function, to respond to soil environment changes to regulate the symplastic growth of aboveground and underground plant sections.
In summary, our findings revealed that RCN1 integrates photoperiodic and drought stress information to delay flowering transition in rice. However, the mechanism of RCN1 action is still unclear, and therefore, requires further detailed investigations.
Materials and Methods
Vector Construction
We used the O. sativa japonica cultivar (cv.) Nipponbare as the genetic background to generate the transgenic rice plants. To examine the expression pattern of RCN1, its promoter (4 kb upstream of ATG) was first amplified by PCR using the genomic DNA of rice, and then inserted into the pCAMIBA1300 vector, which contained the GUS reporter group and the NOS terminator, to generate the pRCN1::GUS:NOS construct. A similar method was used to generate the construct containing GFP. The full length RCN1 coding region was amplified from rice by PCR, and then cloned into the region between the RCN1 promoter and GFP in pRCN1::GUS:NOS to obtain the pRCN1::RCN1-GFP:NOS construct. To generate the DEX-inducible overexpression construct of OsAREB1 and OSBZ8, we amplified the full-length cDNAs of OsAREB1 and OSBZ8 from the genomic DNA by PCR. Fragments of OsAREB1 and OSBZ8 were inserted into the pINDEX vector containing the Gos2 promoter, which can be induced by low concentrations (1–10 μM) of DEX (Ouwerkerk et al. 2001). The CRISPR-Cas9 procedure was performed as previously described (Mao et al. 2013). Two different single-guide RNA oligos targeting RCN1 were designed based on the first and second exons and transformed into Nipponbare. The rcn1–4 and rcn1–11 mutants were used in this study (Figure S1). The primers used in vector construction are shown in Table S1.
Plant Growth Conditions and Drought Stress Experiment
The wild-type and rcn1 mutant plants (two weeks after germination) were grown during the growing season. Flowering time was measured as the number of days from germination to heading. For the drought stress experiments, the plants were grown on nutrient soil in cylindrical boxes (diameter = 20 cm, height = 21 cm). After two weeks, half of the plants were subject to drought stress. Each cylindrical box was filled with nutrient soil and air-dried for 72 h in an oven at 75 °C and weighed to determine dry weight. The cylindrical boxes were then immersed in water for 24 h and the wet weight was determined. The soil water capacity (SWC) was calculated as the difference between wet weight (full water) and dry weight. The soil relative water content (SRWC) was calculated as: (current wet weight - dry weight) / SWC × 100. When rice leaves started to curl, the SRWC was 50%. Fifteen plants were tested for each genotype in both control and drought-treated groups. The cylindrical boxes were weighed every day; nutrient solution was added to maintain the SRWC at 30%–50% (severe drought stress). Sufficient water was supplied to the control group, and both treatment and control groups were supplied with sufficient nutrient solution. The first fully expanded leaf was sampled in at 50 DAS for transcript level analysis under normally watered/drought stress conditions.
Plant Culture and Treatments
Rice cv. Wuyujing3 seeds were sown individually in 96-well plates. The plates were then floated on rectangular plastic trays (32.5 cm × 21.5 cm × 11.5 cm) filled with 4 L of modified Yoshida nutrient solution containing 1.0 mM NH4NO3, 1.0 mM CaCl2, 1.7 mM MgSO4, 0.5 mM K2SO4, 320.0 μM NaH2PO4, 36.0 μM Fe (III)-EDTA, 18.0 μM H3BO3, 9.1 μM MnCl2, 0.15 μM ZnSO4, 0.16 μM CuSO4, 0.074 μM (NH4)6Mo7O24, and 40.5 μM citric acid (Ding et al. 2018). The nutrient solution was refreshed every five days to maintain a pH of 5.5. The plants were grown in climate chambers under LD conditions (14 h light at 28 °C /10 h dark at 24 °C) with a relative humidity of ~ 70%, and treated for 14 days.
The effect of water stress on RCN1 expression was studied using PEG6000 dissolved in nutrient solution at 10%, 20%, or 30% concentration. Plants grown in the nutrient solution without PEG6000 were used as the controls. Root and shoot samples were collected after 24 h of treatment and frozen in liquid nitrogen at − 80 °C.
Plants were treated with ABA at different concentrations and for different times. The stock solution of ABA was dissolved in ethanol and diluted in modified Yoshida nutrient solution. Rice plants were treated with 10 μM ABA; root and shoot samples were collected after 0 (control), 3, 6, 24, 30, 48, 54, 96, and 102 h, and frozen in liquid nitrogen at − 80 °C. Five concentrations of ABA (0.4, 2, 10, 50, and 250 μM) were used, plus a control treatment with an equal amount of the solvent. Root and shoot samples were collected after 24 h of ABA treatment and frozen in liquid nitrogen at − 80 °C.
As the main hormone of the drought response pathway, ABA may play a key role in regulating the response of RCN1 to drought. To verify this hypothesis, further experiments were conducted to identify whether PEG6000 induces RCN1 expression in an ABA-dependent manner. 48 rice plants were treated with fluridone (40 μΜ) for 30 min, and then moved into 30% PEG6000 solution containing fluridone. Thirty rice plants were also treated with 50 mM CHX for 30 min to block protein synthesis, and 30 plants did not receive the treatment. The CHX-treated and untreated rice plants were then divided into two groups, each treated with the nutrient solution or 50 μM ABA. In the ABA-signaling pathway, the released SNF1-related protein kinase 2 activates downstream transcription factors by protein phosphorylation, and then promotes the expression of ABA-response genes (Kerr et al. 2018). Thus, 60 rice plants were pretreated with K252a (50 nM), which is a phosphorylation blocker, while 60 plants did not receive the treatment. The K252a-treated and untreated rice plants were then divided into two groups, and each of these plants was further treated with 30% PEG6000 or 50 μM ABA.
Twenty plants each of Gos2::OsAREB1, Gos2::OSBZ8, and Nipponbare lines, were treated with DEX powder dissolved in ethanol and diluted to 10 μΜ in modified Yoshida nutrient solution for 24 h, while 20 plants were treated with an equal amount of the solvent and used as the controls. Forty plants each of pRCN1::GUS:NOS and pRCN1::RCN1-GFP:NOS lines were grown in water culture under LD condition (14 h light at 28 °C /10 h dark at 24 °C). Half of the plants were treated with 50 μM ABA for 6 h, while the other half was left untreated and used as the controls.
RNA Extraction and qRT-PCR Analysis
The total RNA was extracted from rice tissues with using the E.Z.N.A.® Total RNA Kit I (Omega Bio-tek, Norcross, GA, USA) following the manufacturer’s instructions. The total RNA samples were treated with RQ1 DNase (Promega, Madison, WI, USA) at 37 °C for 30 min to remove genomic DNA, and then converted to cDNA with iScript™ Reverse Transcription Supermix (Bio-Rad, Hercules, CA, USA). A qRT-PCR was conducted in the CFX Connect Real Time System (Bio-Rad) using the iTaq Universal SYBR Green Supermix (Bio-Rad). The thermal cycle conditions were as follows: 95 °C for 3 min and 40 cycles at 95 °C for 15 s and 60 °C for 60 s, followed by melting curve analysis to verify the specificity of amplification. The ΔΔCt method was used to calculate relative expression levels using the rice Actin gene as the internal control (Arocho et al. 2006). The primers used in the qRT-PCR are shown in Table S2. The values presented are the mean of three biological replicates, each of which had three technical replicates.
Histochemical Analysis of GUS and Fluorescence Imaging
GUS staining was performed as previously reported (Tamaki et al. 2007). The GFP fluorescence was detected using the LSM710 META confocal laser-scanning microscope (Carl Zeiss AG, Oberkochen, Germany) as previously reported (Tamaki et al. 2007).
Transient Expression of Proteins in Nicotiana benthamiana Leaf Epidermal Cells
The 35S::RCN1-GFP::NOS vector was acquired from BioRun (Shanghai, P. R. China) and the full length RCN1 coding region was inserted into the vector that contained the GFP reporter group and 35S promoter. The vector was transformed into tobacco (N. benthamiana) leaves using Agrobacterium tumefaciens strain EHA105 (Lee et al. 2005). The GFP fluorescence was observed under a confocal laser microscope (LSM710) after preparation of microscope slides with tobacco leaves at 5 days post-infection.
Protein Expression
The OSBZ8, OsAREB1, and TRAB1 coding regions were cloned into the pCold™ vector (Code No. 3361; Takara Bio Inc., Kusatsu, Japan) and the corresponding proteins were expressed in Escherichia coli BL21 strain according to the manufacturer’s instructions. Collected cells were resuspended in phosphate buffer saline (NaCl 137 mM, KCl 2.7 mM, Na2HPO4 10 mM, KH2PO4 2 mM; pH = 7.2) containing the cOmplete™ Protease Inhibitor Cocktail (Roche, Basel, Switzerland) and lysed with ultrasound. The suspension was centrifuged at 4000×g for 10 min at 4 °C, and the resulting supernatant was used for further experiments. The lysates were purified using the Capturem™ His-Tagged Purification Miniprep Kit (Takara Bio Inc., Kusatsu, Japan) according to the manufacturer’s instructions.
EMSA
The EMSA experiments were performed on the 7-kb upstream and 2-kb downstream DNA sequences of RCN1. Eleven DNA fragments containing 19 ACGT were synthesized and labeled with biotin (Table S3). The DNA-binding reaction was performed using the Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions.
Statistical Analyses
The results are presented as mean ± standard error. The data were analyzed using a two-tailed paired Student’s t test.
Availability of Data and Materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
Abbreviations
- ABA:
-
Abscisic acid
- CHX:
-
Cycloheximide
- CRISPR-Cas9:
-
Clustered regularly interspaced short palindromic repeats-associated protein-9
- DE:
-
Drought-escape
- DEX:
-
Dexamethasone
- EMSA:
-
Electrophoretic mobility shift assays
- GFP:
-
GREEN FLUORESCENT PROTEIN
- GUS:
-
Beta-glucuronidase
- LD:
-
Long-day
- PEG6000:
-
Polyethylene glycol
- qRT-PCR:
-
Quantitative reverse transcription- PCR
- SAM:
-
Shoot apical meristem
- SRWC:
-
Soil relative water content
- SWC:
-
Soil water capacity
- WT:
-
Wild type
References
Arocho A, Chen BY, Ladanyi M, Pan QL (2006) Validation of the 2(−Delta Delta Ct) calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn Mol Pathol 15:56–61
Bernal M, Estiarte M, Penuelas J (2011) Drought advances spring growth phenology of the Mediterranean shrub Erica multiflora. Plant Biol (Stuttg) 13:252–257
Bocco R, Lorieux M, Seck PA, Futakuchi K, Manneh B, Baimey H, Ndjiondjop MN (2012) Agro-morphological characterization of a population of introgression lines derived from crosses between IR 64 (Oryza sativa indica) and TOG 5681 (Oryza glaberrima) for drought tolerance. Plant Sci 183:65–76
Brambilla V, Martignago D, Goretti D, Cerise M, Somssich M, de Rosa M, Galbiati F, Shrestha R, Lazzaro F, Simon R, Fornara F (2017) Antagonistic transcription factor complexes modulate the floral transition in rice. Plant Cell 29:2801–2816
Corbesier L et al (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316:1030–1033
Ding CQ, Chang ZY, Wang Y, You SL, Wang SH, Ding YF (2018) Proteomic analysis reveals that developing leaves are more sensitive to nitrogen fertilizer than mature leaves. J Plant Growth Regul 37:426–437
Dixit S, Singh A, Kumar A (2014) Rice breeding for high grain yield under drought: a strategic solution to a complex problem. Int J Agronomy 2014:1–15
Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18:926–936
Du H, Huang F, Wu N, Li X, Hu H, Xiong L (2018) Integrative regulation of drought escape through ABA-dependent and -independent pathways in rice. Mol Plant 11:584–597
Franks SJ (2011) Plasticity and evolution in drought avoidance and escape in the annual plant Brassica rapa. New Phytol 190:249–257
Fukai S, Pantuwan G, Jongdee B, Cooper M (1999) Screening for drought resistance in rainfed lowland rice. Field Crops Res 64:61–74
Galbiati F, Chiozzotto R, Locatelli F, Spada A, Genga A, Fornara F (2016) Hd3a, RFT1 and Ehd1 integrate photoperiodic and drought stress signals to delay the floral transition in rice. Plant Cell Environ 39:1982–1993
Guo Y, Hans H, Christian J, Molina C (2014) Mutations in single FT- and TFL1-paralogs of rapeseed (Brassica napus L.) and their impact on flowering time and yield components. Front Plant Sci 5:282
Hanano S, Goto K (2011) Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23:3172–3184
Hattori T, Totsuka M, Hobo T, Kagaya Y, Yamamoto-Toyoda A (2002) Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol 43:136–140
Hoang GT et al (2019) Genome-wide association study of a panel of vietnamese rice landraces reveals new QTLs for tolerance to water deficit during the vegetative phase. Rice (N Y) 12:4
HSU YT, KAO CH (2003) Role of abscisic acid in cadmium tolerance of rice (Oryza sativa L.) seedlings. Plant Cell Environ 26:867–874
Hsu YT, Kuo MC, Kao CH (2006) Cadmium-induced ammonium ion accumulation of rice seedlings at high temperature is mediated through abscisic acid. Plant Soil 287:267–277
Huang X et al (2012) Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet 44:32–39
Jin XF, Xiong AS, Peng RH, Liu JG, Gao F, Chen JM, Yao QH (2010) OsAREB1, an ABRE-binding protein responding to ABA and glucose, has multiple functions in Arabidopsis. BMB Rep 43:34–39
Kaneko-Suzuki M et al (2018) TFL1-like proteins in rice antagonize rice FT-like protein in inflorescence development by competition for complex formation with 14-3-3 and FD. Plant Cell Physiol 59:458–468
Kerr TCC, Abdel-Mageed H, Aleman L, Lee J, Payton P, Cryer D, Allen RD (2018) Ectopic expression of two AREB/ABF orthologs increases drought tolerance in cotton (Gossypium hirsutum). Plant Cell Environ 41:898–907
Kowalczyk-Schroder S, Sandmann G (1992) Interaction of fluridone with phytoene desaturation of Aphanocapsa. Pestic Biochem Physiol 42:7–12
Lee JY, Taoka K, Yoo BC, Ben-Nissan G, Kim DJ, Lucas WJ (2005) Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell 17:2817–2831
Liu L et al (2012) FTIP1 is an essential regulator required for florigen transport. PLoS Biol 10:e1001313
Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK (2013) Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant 6:2008–2011
Nakagawa M, Shimamoto K, Kyozuka J (2002) Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J 29:743–750
Nakashima K, Yamaguchi-Shinozaki K (2013) ABA signaling in stress-response and seed development. Plant Cell Rep 32:959–970
Ouwerkerk PB, de Kam RJ, Hoge JH, Meijer AH (2001) Glucocorticoid-inducible gene expression in rice. Planta 213:370–378
Perales L, Arbona V, Gomez-Cadenas A, Cornejo MJ, Sanz A (2005) A relationship between tolerance to dehydration of rice cell lines and ability for ABA synthesis under stress. Plant Physiol Biochem 43:786–792
Raikhel NV, Palevitz BA, Haigler CH (1986) Abscisic acid control of lectin accumulation in wheat seedlings and callus cultures : effects of exogenous ABA and fluridone. Plant Physiol 80:167–171
Riboni M, Galbiati M, Tonelli C, Conti L (2013) GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS. Plant Physiol 162:1706–1719
Riboni M, Robustelli Test A, Galbiati M, Tonelli C, Conti L (2016) ABA-dependent control of GIGANTEA signalling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J Exp Bot 67:6309–6322
Sherrard ME, Maherali H (2006) The adaptive significance of drought escape in Avena barbata, an annual grass. Evolution 60:2478–2489
Shi B, Ni L, Zhang AY, Cao JM, Zhang H, Qin TT, Tan MP, Zhang JH, Jiang MY (2012) OsDMI3 is a novel component of abscisic acid signaling in the induction of antioxidant defense in leaves of rice. Mol Plant 5:1359–1374
Song S et al (2017) OsFTIP1-mediated regulation of florigen transport in rice is negatively regulated by the ubiquitin-like domain kinase OsUbDKgamma4. Plant Cell 29:491–507
Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316:1033–1036
Taoka K et al (2011) 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476:332–335
Wu CY, You CJ, Li CS, Long T, Chen GX, Byrne ME, Zhang QF (2008) RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc Natl Acad Sci U S A 105:12915–12920
Wu JJ, He B, Lu AF, Zhou L, Liu M, Zhao L (2011) Quantitative assessment and spatial characteristics analysis of agricultural drought vulnerability in China. Nat Hazards 56:785–801
Zhang CY, Liu J, Zhao T, Gomez A, Li C, Yu CS, Li HY, Lin JZ, Yang YZ, Liu B, Lin CT (2016) A drought-inducible transcription factor delays reproductive timing in rice. Plant Physiol 171(1):334–343
Zhang SH, Hu WJ, Wang LP, Lin CF, Cong B, Sun CR, Luo D (2005) TFL1/CEN-like genes control intercalary meristem activity and phase transition in rice. Plant Sci 168:1393–1408
Zhang Y, Luo L, Xu C, Zhang Q, Xing Y (2006) Quantitative trait loci for panicle size, heading date and plant height co-segregating in trait-performance derived near-isogenic lines of rice (Oryza sativa). Theor Appl Genet 113:361–368
Acknowledgments
We thank Jian-Kang Zhu for providing the vectors of the CRISPR-Cas9 system.
Funding
This study was supported by the National Natural Science Foundation of China (Grant number 31872855), Research Foundation of Education Bureau of Hunan Province, China (Grant number 18C0578), and the Fundamental Research Funds for the Central Universities (Grant Numbers KYZ201803).
Author information
Authors and Affiliations
Contributions
C.D., Y.W., Y.L., and Y.D. designed the research. Y.W., Y.L., C.D., and Z.G. performed the experiments. C.D. and Y.W. analyzed the data and prepared the manuscript. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
Mutational sites of RCN1 in rcn1–4 and rcn1–11 mutants. Figure S2. Schematic diagram of the experimental timetable for drought treatment. SRWC: soil relative water content. Figure S3. The expression levels of RCN1 (a), Hd3a (b), and RFT1 (c) in the leaves response to drought. Figure S4. Localization of the RCN1-GFP protein in tobacco leaf epidermal cells (a) and rice protoplasts (b). Figure S5. Expression levels of bZIP family transcription factors under ABA treatment in rice roots, according to the RiceXpro data (a). Expression levels of OsAREB1, TRAB1, OSBZ8, RITA, bZIP23, and RCN1 in rice roots (b) and leaves (c) under ABA treatment. Expression levels of OsAREB1, OSBZ8, and TRAB1 in rice roots (d) and leaves (e) after CHX and ABA treatments. CHX: cycloheximide, a type of protein de novo synthesis blocker. Figure S6. Gel electrophoresis revealed that OsAREB1 (a) and OSBZ8 (b) were not bound to the upstream sequence of RCN1; TRAB1 was not bound to the sequence of RCN1 (c). Figure S7. The expression levels of OsAREB1 (a), OSBZ8 (b) in the leaves response to drought at the timing of flowering. Table S1. Primers used for vector construction. Table S2. Primers used for qRT-PCR. Table S3. Probes used for EMSA experiment.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, Y., Lu, Y., Guo, Z. et al. RICE CENTRORADIALIS 1, a TFL1-like Gene, Responses to Drought Stress and Regulates Rice Flowering Transition. Rice 13, 70 (2020). https://doi.org/10.1186/s12284-020-00430-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-020-00430-3